|
|
|
|
|
| Sample: |
Bruton's tyrosine kinase - Src homology 3-2 kinase domain monomer, 52 kDa Homo sapiens protein
|
| Buffer: |
20mM Tris, 150mM NaCl, 1mM TCEP, 5% glycerol, pH: 7.5
|
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Nov 2
|
Btk SH2-kinase interface is critical for allosteric kinase activation and its targeting inhibits B-cell neoplasms
Nature Communications 11(1) (2020)
Duarte D, Lamontanara A, La Sala G, Jeong S, Sohn Y, Panjkovich A, Georgeon S, Kükenshöner T, Marcaida M, Pojer F, De Vivo M, Svergun D, Kim H, Dal Peraro M, Hantschel O
|
| RgGuinier |
2.6 |
nm |
| Dmax |
8.3 |
nm |
| VolumePorod |
72 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Bruton's tyrosine kinase - full length monomer, 77 kDa Homo sapiens protein
|
| Buffer: |
20mM Tris, 150mM NaCl, 1mM TCEP, 5% glycerol, pH: 7.5
|
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Nov 3
|
Btk SH2-kinase interface is critical for allosteric kinase activation and its targeting inhibits B-cell neoplasms
Nature Communications 11(1) (2020)
Duarte D, Lamontanara A, La Sala G, Jeong S, Sohn Y, Panjkovich A, Georgeon S, Kükenshöner T, Marcaida M, Pojer F, De Vivo M, Svergun D, Kim H, Dal Peraro M, Hantschel O
|
| RgGuinier |
4.0 |
nm |
| Dmax |
15.6 |
nm |
| VolumePorod |
114 |
nm3 |
|
|
|
|
|
|
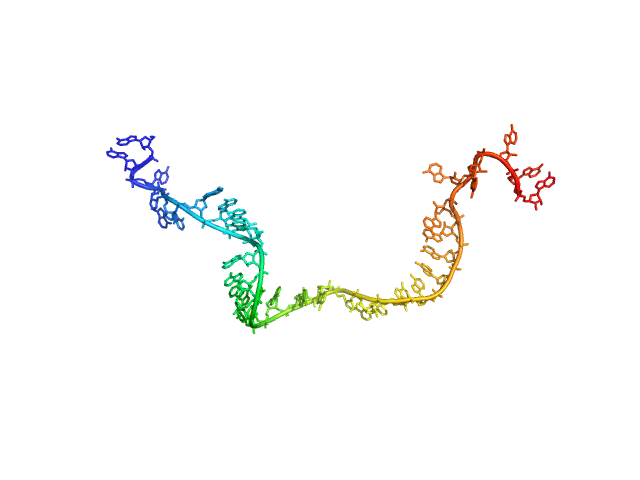
|
| Sample: |
Poly-adenosine monomer, 10 kDa RNA
|
| Buffer: |
1 mM Na-MOPS, 20 mM NaCl, 20 µM EDTA, pH: 7
|
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2015 Oct 24
|
Visualizing disordered single-stranded RNA: connecting sequence, structure and electrostatics.
J Am Chem Soc (2019)
Plumridge A, Andresen K, Pollack L
|
| RgGuinier |
2.7 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
17 |
nm3 |
|
|
|
|
|
|
|
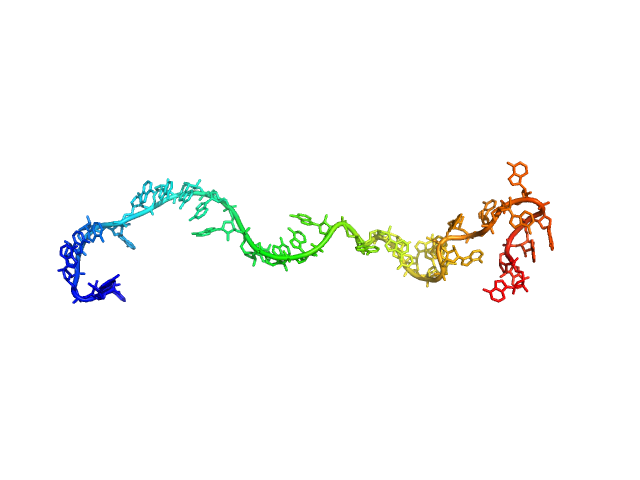
| Sample: |
Poly-adenosine monomer, 10 kDa RNA
|
| Buffer: |
1 mM Na-MOPS, 100 mM NaCl, 20 µM EDTA, pH: 7
|
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2015 Oct 25
|
Visualizing disordered single-stranded RNA: connecting sequence, structure and electrostatics.
J Am Chem Soc (2019)
Plumridge A, Andresen K, Pollack L
|
| RgGuinier |
2.4 |
nm |
| Dmax |
10.7 |
nm |
| VolumePorod |
14 |
nm3 |
|
|
|
|
|
|
|
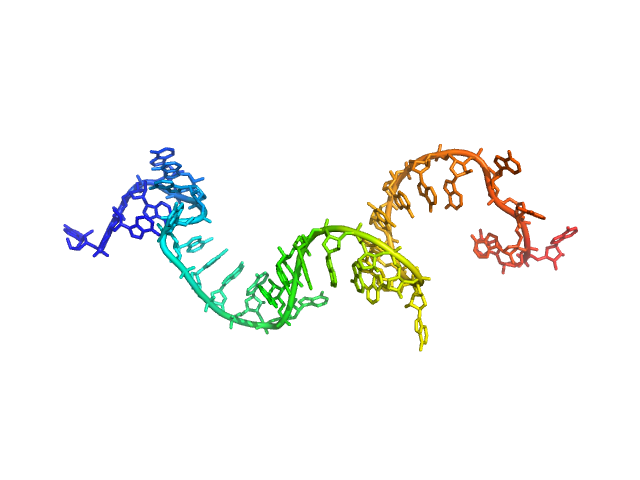
| Sample: |
Poly-adenosine monomer, 10 kDa RNA
|
| Buffer: |
1 mM Na-MOPS, 200 mM NaCl, 20 µM EDTA, pH: 7
|
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2015 Oct 25
|
Visualizing disordered single-stranded RNA: connecting sequence, structure and electrostatics.
J Am Chem Soc (2019)
Plumridge A, Andresen K, Pollack L
|
| RgGuinier |
2.5 |
nm |
| Dmax |
10.5 |
nm |
| VolumePorod |
14 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Poly-adenosine monomer, 10 kDa RNA
|
| Buffer: |
1 mM Na-MOPS, 400 mM NaCl, 20 µM EDTA, pH: 7
|
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2015 Oct 25
|
Visualizing disordered single-stranded RNA: connecting sequence, structure and electrostatics.
J Am Chem Soc (2019)
Plumridge A, Andresen K, Pollack L
|
| RgGuinier |
2.4 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
13 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Poly-adenosine monomer, 10 kDa RNA
|
| Buffer: |
1 mM Na-MOPS, 600 mM NaCl, 20 µM EDTA, pH: 7
|
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2015 Oct 25
|
Visualizing disordered single-stranded RNA: connecting sequence, structure and electrostatics.
J Am Chem Soc (2019)
Plumridge A, Andresen K, Pollack L
|
| RgGuinier |
2.2 |
nm |
| Dmax |
10.2 |
nm |
| VolumePorod |
13 |
nm3 |
|
|
|
|
|
|
|
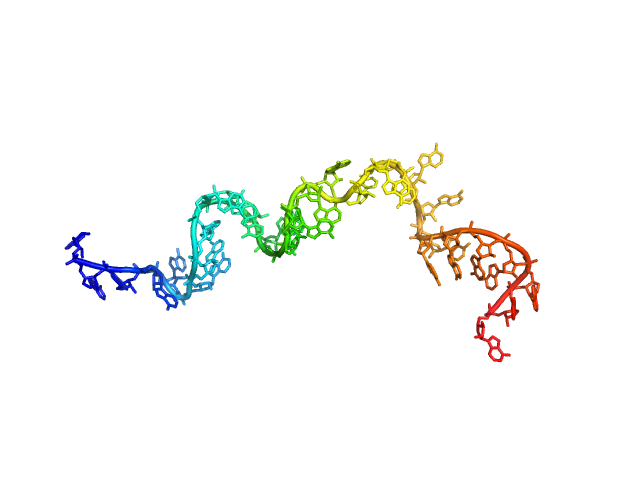
| Sample: |
Poly-adenosine monomer, 10 kDa RNA
|
| Buffer: |
1 mM Na-MOPS, 20 mM NaCl, 1 mM MgCl2, 20 µM EDTA, pH: 7
|
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2019 Jul 24
|
Visualizing disordered single-stranded RNA: connecting sequence, structure and electrostatics.
J Am Chem Soc (2019)
Plumridge A, Andresen K, Pollack L
|
| RgGuinier |
2.5 |
nm |
| Dmax |
10.4 |
nm |
| VolumePorod |
12 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Poly-adenosine monomer, 10 kDa RNA
|
| Buffer: |
1 mM Na-MOPS, 20 mM NaCl, 2 mM MgCl2, 20 µM EDTA, pH: 7
|
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2015 Oct 24
|
Visualizing disordered single-stranded RNA: connecting sequence, structure and electrostatics.
J Am Chem Soc (2019)
Plumridge A, Andresen K, Pollack L
|
| RgGuinier |
2.4 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
12 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Poly-adenosine monomer, 10 kDa RNA
|
| Buffer: |
1 mM Na-MOPS, 20 mM NaCl, 5 mM MgCl2, 20µM EDTA, pH: 7
|
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2015 Oct 24
|
Visualizing disordered single-stranded RNA: connecting sequence, structure and electrostatics.
J Am Chem Soc (2019)
Plumridge A, Andresen K, Pollack L
|
| RgGuinier |
2.3 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
15 |
nm3 |
|
|