|
|
|
|
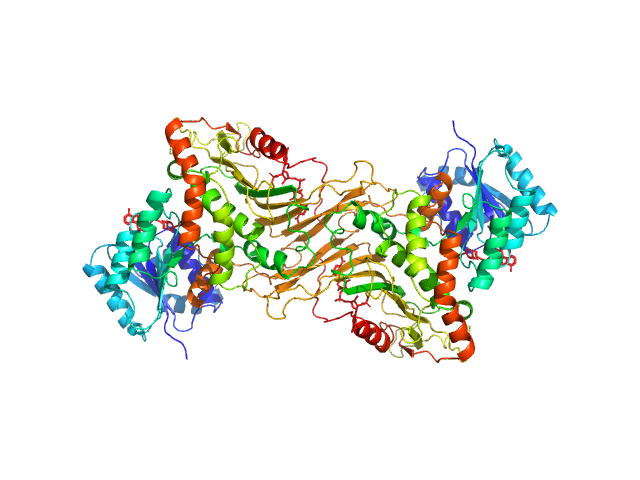
|
| Sample: |
Glucose-6-phosphate 1-dehydrogenase dimer, 119 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, pH: 8
|
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2019 Jul 24
|
Long-range structural defects by pathogenic mutations in most severe glucose-6-phosphate dehydrogenase deficiency
Proceedings of the National Academy of Sciences 118(4) (2021)
Horikoshi N, Hwang S, Gati C, Matsui T, Castillo-Orellana C, Raub A, Garcia A, Jabbarpour F, Batyuk A, Broweleit J, Xiang X, Chiang A, Broweleit R, Vöhringer-Martinez E, Mochly-Rosen D, Wakatsuki S
|
| RgGuinier |
3.6 |
nm |
| Dmax |
12.1 |
nm |
| VolumePorod |
160 |
nm3 |
|
|
|
|
|
|
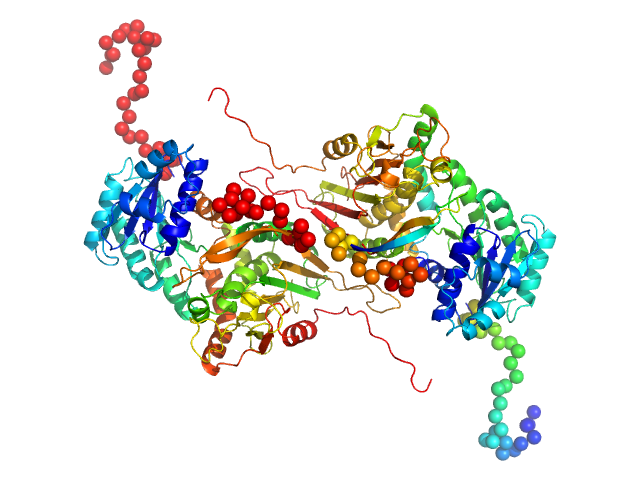
|
| Sample: |
Glucose-6-phosphate 1-dehydrogenase P396L dimer, 119 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, pH: 8
|
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2019 Jul 24
|
Long-range structural defects by pathogenic mutations in most severe glucose-6-phosphate dehydrogenase deficiency
Proceedings of the National Academy of Sciences 118(4) (2021)
Horikoshi N, Hwang S, Gati C, Matsui T, Castillo-Orellana C, Raub A, Garcia A, Jabbarpour F, Batyuk A, Broweleit J, Xiang X, Chiang A, Broweleit R, Vöhringer-Martinez E, Mochly-Rosen D, Wakatsuki S
|
| RgGuinier |
3.7 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
178 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Di-domain acyl carrier protein of PigH from prodigiosin biosynthesis monomer, 22 kDa Serratia sp. ATCC … protein
|
| Buffer: |
20 mM Tris supplemented with 5 mM DTT, pH: 7
|
| Experiment: |
SAXS
data collected at BL1.3W, Synchrotron Light Research Institute (SLRI) on 2016 Jan 13
|
Solution Structure and Conformational Dynamics of a Doublet Acyl Carrier Protein from Prodigiosin Biosynthesis.
Biochemistry (2021)
Thongkawphueak T, Winter AJ, Williams C, Maple HJ, Soontaranon S, Kaewhan C, Campopiano DJ, Crump MP, Wattana-Amorn P
|
| RgGuinier |
2.4 |
nm |
| Dmax |
8.4 |
nm |
| VolumePorod |
39 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
4-hydroxy-2,2'-bipyrrole-5-methanol synthase PigH monomer, 22 kDa Serratia sp. protein
|
| Buffer: |
25 mM Tris, 150 mM NaCl, pH: 7.5
|
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2020 Mar 3
|
Solution Structure and Conformational Dynamics of a Doublet Acyl Carrier Protein from Prodigiosin Biosynthesis.
Biochemistry (2021)
Thongkawphueak T, Winter AJ, Williams C, Maple HJ, Soontaranon S, Kaewhan C, Campopiano DJ, Crump MP, Wattana-Amorn P
|
| RgGuinier |
2.9 |
nm |
| Dmax |
9.9 |
nm |
| VolumePorod |
83 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
4-hydroxy-2,2'-bipyrrole-5-methanol synthase PigH monomer, 22 kDa Serratia sp. protein
|
| Buffer: |
25 mM Tris, 150 mM NaCl, pH: 7.5
|
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2020 Mar 3
|
Solution Structure and Conformational Dynamics of a Doublet Acyl Carrier Protein from Prodigiosin Biosynthesis.
Biochemistry (2021)
Thongkawphueak T, Winter AJ, Williams C, Maple HJ, Soontaranon S, Kaewhan C, Campopiano DJ, Crump MP, Wattana-Amorn P
|
| RgGuinier |
2.8 |
nm |
| Dmax |
9.7 |
nm |
| VolumePorod |
72 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Apolipoprotein D tetramer, 77 kDa Homo sapiens protein
|
| Buffer: |
50 mM Na Phosphate, 150 mM NaCl, 3% glycerol, pH: 7.4
|
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2016 Nov 11
|
Small angle X-ray scattering analysis of ligand-bound forms of tetrameric apolipoprotein-D
Bioscience Reports 41(1) (2021)
Kielkopf C, Whitten A, Garner B, Brown S
|
| RgGuinier |
3.3 |
nm |
| Dmax |
10.5 |
nm |
| VolumePorod |
168 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Apolipoprotein D tetramer, 77 kDa Homo sapiens protein
|
| Buffer: |
50 mM Na Phosphate, 150 mM NaCl, 3% glycerol, pH: 7.4
|
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2016 Nov 11
|
Small angle X-ray scattering analysis of ligand-bound forms of tetrameric apolipoprotein-D
Bioscience Reports 41(1) (2021)
Kielkopf C, Whitten A, Garner B, Brown S
|
| RgGuinier |
3.3 |
nm |
| Dmax |
10.3 |
nm |
| VolumePorod |
164 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Apolipoprotein D tetramer, 77 kDa Homo sapiens protein
|
| Buffer: |
50 mM Na Phosphate, 150 mM NaCl, 3% glycerol, pH: 7.4
|
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2016 Nov 11
|
Small angle X-ray scattering analysis of ligand-bound forms of tetrameric apolipoprotein-D
Bioscience Reports 41(1) (2021)
Kielkopf C, Whitten A, Garner B, Brown S
|
| RgGuinier |
3.3 |
nm |
| Dmax |
10.6 |
nm |
| VolumePorod |
163 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Apolipoprotein D tetramer, 77 kDa Homo sapiens protein
|
| Buffer: |
50 mM Na Phosphate, 150 mM NaCl, 3% glycerol, pH: 7.4
|
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2016 Nov 11
|
Small angle X-ray scattering analysis of ligand-bound forms of tetrameric apolipoprotein-D
Bioscience Reports 41(1) (2021)
Kielkopf C, Whitten A, Garner B, Brown S
|
| RgGuinier |
3.3 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
161 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Apolipoprotein D tetramer, 77 kDa Homo sapiens protein
|
| Buffer: |
50 mM Na Phosphate, 150 mM NaCl, 3% glycerol, pH: 7.4
|
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2016 Nov 11
|
Small angle X-ray scattering analysis of ligand-bound forms of tetrameric apolipoprotein-D
Bioscience Reports 41(1) (2021)
Kielkopf C, Whitten A, Garner B, Brown S
|
| RgGuinier |
3.3 |
nm |
| Dmax |
9.9 |
nm |
| VolumePorod |
171 |
nm3 |
|
|