|
|
|
|
|
| Sample: |
Two titin I27 domains linked with RS linker monomer, 23 kDa protein
|
| Buffer: |
50 mM potassium phosphate, 300 mM KCl, DTT 2mM, pH: 7.6
|
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Dec 4
|
Interdomain linkers tailor the stability of immunoglobulin repeats in polyproteins.
Biochem Biophys Res Commun 550:43-48 (2021)
Joshi T, Garg S, Estaña A, Cortés J, Bernadó P, Das S, Kammath AR, Sagar A, Rakshit S
|
|
|
|
|
|
|
|
| Sample: |
Two titin I27 domains linked with RS linker monomer, 23 kDa protein
|
| Buffer: |
50 mM potassium phosphate, 300 mM KCl, DTT 2mM, pH: 7.6
|
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Dec 4
|
Interdomain linkers tailor the stability of immunoglobulin repeats in polyproteins.
Biochem Biophys Res Commun 550:43-48 (2021)
Joshi T, Garg S, Estaña A, Cortés J, Bernadó P, Das S, Kammath AR, Sagar A, Rakshit S
|
|
|
|
|
|
|
|
| Sample: |
Two titin I27 domains linked with GGG linker monomer, 23 kDa protein
|
| Buffer: |
50 mM potassium phosphate, 300 mM KCl, DTT 2mM, pH: 7.6
|
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Dec 4
|
Interdomain linkers tailor the stability of immunoglobulin repeats in polyproteins.
Biochem Biophys Res Commun 550:43-48 (2021)
Joshi T, Garg S, Estaña A, Cortés J, Bernadó P, Das S, Kammath AR, Sagar A, Rakshit S
|
|
|
|
|
|
|
|
| Sample: |
Two titin I27 domains linked with VKA linker monomer, 24 kDa protein
|
| Buffer: |
50 mM potassium phosphate, 300 mM KCl, DTT 2mM, pH: 7.6
|
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Dec 4
|
Interdomain linkers tailor the stability of immunoglobulin repeats in polyproteins.
Biochem Biophys Res Commun 550:43-48 (2021)
Joshi T, Garg S, Estaña A, Cortés J, Bernadó P, Das S, Kammath AR, Sagar A, Rakshit S
|
|
|
|
|
|
|
|
| Sample: |
Two titin I27 domains linked with AAE linker monomer, 24 kDa protein
|
| Buffer: |
50 mM potassium phosphate, 300 mM KCl, DTT 2mM, pH: 7.6
|
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Dec 4
|
Interdomain linkers tailor the stability of immunoglobulin repeats in polyproteins.
Biochem Biophys Res Commun 550:43-48 (2021)
Joshi T, Garg S, Estaña A, Cortés J, Bernadó P, Das S, Kammath AR, Sagar A, Rakshit S
|
|
|
|
|
|
|
|
| Sample: |
Polymerase delta-interacting protein 2 monomer, 37 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES pH 7.5, 500 mM NaCl, 5% (v/v) glycerol, pH: 7.5
|
| Experiment: |
SAXS
data collected at Bruker Nanostar, University of Huddersfield on 2019 Jul 18
|
Crystal structure and molecular dynamics of human POLDIP2, a multifaceted adaptor protein in metabolism and genome stability.
Protein Sci (2021)
Kulik AA, Maruszczak KK, Thomas DC, Nabi-Aldridge NLA, Carr M, Bingham RJ, Cooper CDO
|
| RgGuinier |
2.2 |
nm |
| Dmax |
7.2 |
nm |
| VolumePorod |
56 |
nm3 |
|
|
|
|
|
|
|
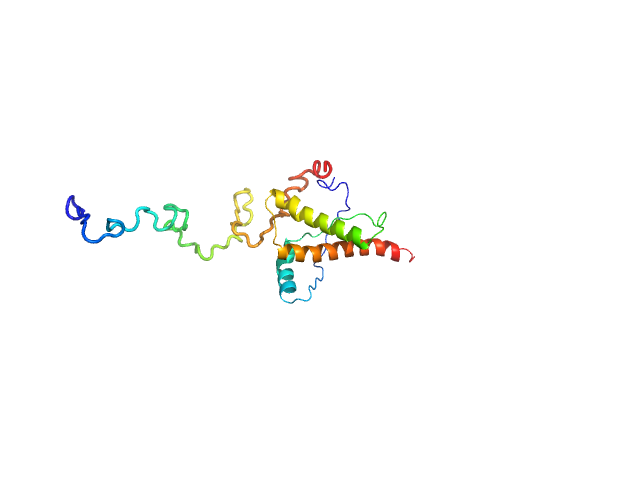
| Sample: |
Major prion protein monomer, 23 kDa Mus musculus protein
|
| Buffer: |
10mM HEPES, 150 mM NaCl, pH: 7.5
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Dec 15
|
Characterization of murine prion protein
Stefano Da Vela
|
| RgGuinier |
2.8 |
nm |
| Dmax |
9.9 |
nm |
| VolumePorod |
43 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Prosaposin (Saposin A) monomer, 10 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris-HCl, 150 mM NaCl, pH: 7.5
|
| Experiment: |
SAXS
data collected at 12-ID-B SAXS/WAXS, Advanced Photon Source (APS), Argonne National Laboratory on 2020 Feb 18
|
Stable Picodisc Assemblies from Saposin Proteins and Branched Detergents.
Biochemistry 60(14):1108-1119 (2021)
Kurgan KW, Chen B, Brown KA, Falco Cobra P, Ye X, Ge Y, Gellman SH
|
| RgGuinier |
1.5 |
nm |
| Dmax |
5.0 |
nm |
| VolumePorod |
23 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Prosaposin (Saposin-A) dimer, 19 kDa Homo sapiens protein
1,2,5,6-tetra-β-D-glucopyranoside-3,4-O-Di-dodecyl-d-mannitol decamer, 12 kDa
|
| Buffer: |
25 mM Tris-HCl, 150 mM NaCl, pH: 7.5
|
| Experiment: |
SAXS
data collected at 12-ID-B SAXS/WAXS, Advanced Photon Source (APS), Argonne National Laboratory on 2020 Feb 18
|
Stable Picodisc Assemblies from Saposin Proteins and Branched Detergents.
Biochemistry 60(14):1108-1119 (2021)
Kurgan KW, Chen B, Brown KA, Falco Cobra P, Ye X, Ge Y, Gellman SH
|
| RgGuinier |
2.3 |
nm |
| Dmax |
6.2 |
nm |
|
|
|
|
|
|
|
| Sample: |
Prosaposin (Saposin-A) dimer, 19 kDa Homo sapiens protein
1,2,5,6-tetra-β-D-glucopyranoside-3,4-O-Di-tridecyl-d-mannitol decamer, 12 kDa
|
| Buffer: |
25 mM Tris-HCl, 150 mM NaCl, pH: 7.5
|
| Experiment: |
SAXS
data collected at 12-ID-B SAXS/WAXS, Advanced Photon Source (APS), Argonne National Laboratory on 2020 Feb 18
|
Stable Picodisc Assemblies from Saposin Proteins and Branched Detergents.
Biochemistry 60(14):1108-1119 (2021)
Kurgan KW, Chen B, Brown KA, Falco Cobra P, Ye X, Ge Y, Gellman SH
|
| RgGuinier |
2.5 |
nm |
| Dmax |
6.7 |
nm |
|
|