|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDKB4_fit1_model1.png)
|
| Sample: |
Properdin (trimer) trimer, 165 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 7.5
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Nov 14
|
Properdin oligomers adopt rigid extended conformations supporting function.
Elife 10 (2021)
Pedersen DV, Pedersen MN, Mazarakis SM, Wang Y, Lindorff-Larsen K, Arleth L, Andersen GR
|
| RgGuinier |
10.2 |
nm |
| Dmax |
27.0 |
nm |
|
|
|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDKC4_fit1_model1.png)
|
| Sample: |
Properdin (tetramer) tetramer, 220 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 7.5
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Nov 14
|
Properdin oligomers adopt rigid extended conformations supporting function.
Elife 10 (2021)
Pedersen DV, Pedersen MN, Mazarakis SM, Wang Y, Lindorff-Larsen K, Arleth L, Andersen GR
|
| RgGuinier |
13.1 |
nm |
| Dmax |
36.0 |
nm |
|
|
|
|
|
|
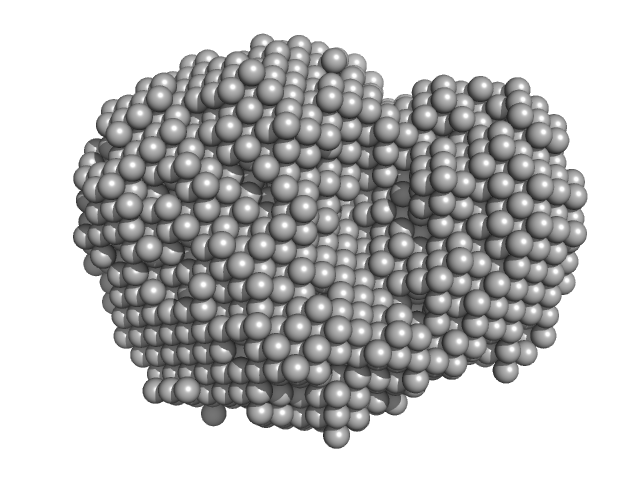
|
| Sample: |
Human telomere G-quadruplex hybrid-2 form monomer, 8 kDa synthetic construct DNA
|
| Buffer: |
6 mM Na2HPO4, 2 mM NaH2PO4, 1 mM Na2EDTA, 185 mM KCl, pH: 7.2
|
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2019 Aug 14
|
The solution structures of higher-order human telomere G-quadruplex multimers.
Nucleic Acids Res (2021)
Monsen RC, Chakravarthy S, Dean WL, Chaires JB, Trent JO
|
| RgGuinier |
1.2 |
nm |
| Dmax |
3.8 |
nm |
| VolumePorod |
9 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Human Telomere Repeat (TTAGGG)8 monomer, 15 kDa synthetic construct DNA
|
| Buffer: |
6 mM Na2HPO4, 2 mM NaH2PO4, 1 mM Na2EDTA, 185 mM KCl, pH: 7.2
|
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2019 Aug 14
|
The solution structures of higher-order human telomere G-quadruplex multimers.
Nucleic Acids Res (2021)
Monsen RC, Chakravarthy S, Dean WL, Chaires JB, Trent JO
|
| RgGuinier |
1.9 |
nm |
| Dmax |
6.4 |
nm |
| VolumePorod |
16 |
nm3 |
|
|
|
|
|
|
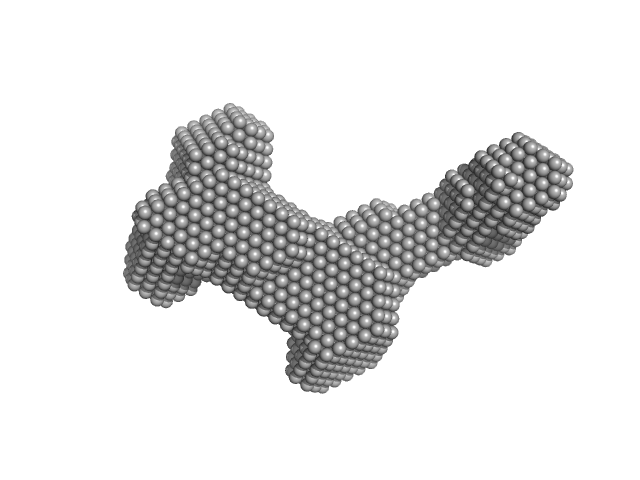
|
| Sample: |
Human Telomere Repeat (TTAGGG)12 monomer, 23 kDa synthetic construct DNA
|
| Buffer: |
6 mM Na2HPO4, 2 mM NaH2PO4, 1 mM Na2EDTA, 185 mM KCl, pH: 7.2
|
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2019 Aug 14
|
The solution structures of higher-order human telomere G-quadruplex multimers.
Nucleic Acids Res (2021)
Monsen RC, Chakravarthy S, Dean WL, Chaires JB, Trent JO
|
| RgGuinier |
2.5 |
nm |
| Dmax |
8.7 |
nm |
| VolumePorod |
26 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Human Telomere 96mer monomer, 31 kDa synthetic construct DNA
|
| Buffer: |
6 mM Na2HPO4, 2 mM NaH2PO4, 1 mM Na2EDTA, 185 mM KCl, pH: 7.2
|
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2020 Feb 19
|
The solution structures of higher-order human telomere G-quadruplex multimers.
Nucleic Acids Res (2021)
Monsen RC, Chakravarthy S, Dean WL, Chaires JB, Trent JO
|
| RgGuinier |
3.2 |
nm |
| Dmax |
10.9 |
nm |
| VolumePorod |
38 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Angiopoietin-like protein 3 (N-terminal) hexamer, 161 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris-HCl pH 7.5, 400 mM NaCl, 2% glycerol, pH: 7.5
|
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 Mar 12
|
Comparison of angiopoietin-like protein 3 and 4 reveals structural and mechanistic similarities.
J Biol Chem :100312 (2021)
Gunn KH, Gutgsell AR, Xu Y, Johnson CV, Liu J, Neher SB
|
| RgGuinier |
5.6 |
nm |
| Dmax |
34.0 |
nm |
| VolumePorod |
380 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Angiopoietin-related protein 3 trimer, 81 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris-HCl pH 7.5, 400 mM NaCl, 2% glycerol, pH: 7.5
|
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 Mar 12
|
Comparison of angiopoietin-like protein 3 and 4 reveals structural and mechanistic similarities.
J Biol Chem :100312 (2021)
Gunn KH, Gutgsell AR, Xu Y, Johnson CV, Liu J, Neher SB
|
| RgGuinier |
4.6 |
nm |
| Dmax |
21.5 |
nm |
| VolumePorod |
71 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Angiopoietin-related protein 4 trimer, 46 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris-HCl pH 7.4, 300 mM NaCl, 100 mM betaine, 500 mM arginine, pH: 7.4
|
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2017 Nov 8
|
Comparison of angiopoietin-like protein 3 and 4 reveals structural and mechanistic similarities.
J Biol Chem :100312 (2021)
Gunn KH, Gutgsell AR, Xu Y, Johnson CV, Liu J, Neher SB
|
| RgGuinier |
4.7 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
69 |
nm3 |
|
|
|
|
|
|
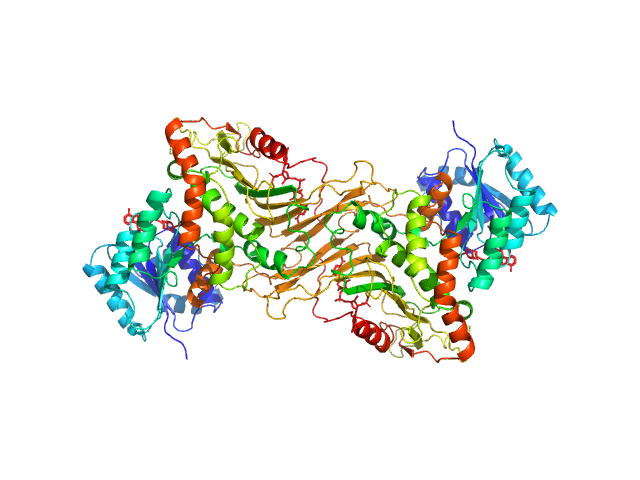
|
| Sample: |
Glucose-6-phosphate 1-dehydrogenase dimer, 119 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, pH: 8
|
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2019 Jul 24
|
Long-range structural defects by pathogenic mutations in most severe glucose-6-phosphate dehydrogenase deficiency
Proceedings of the National Academy of Sciences 118(4) (2021)
Horikoshi N, Hwang S, Gati C, Matsui T, Castillo-Orellana C, Raub A, Garcia A, Jabbarpour F, Batyuk A, Broweleit J, Xiang X, Chiang A, Broweleit R, Vöhringer-Martinez E, Mochly-Rosen D, Wakatsuki S
|
| RgGuinier |
3.6 |
nm |
| Dmax |
12.1 |
nm |
| VolumePorod |
160 |
nm3 |
|
|