|
|
|
|
|
| Sample: |
Menisporopsin singlet acyl carrier protein-thioesterase monomer, 45 kDa Menisporopsin theobromae BCC … protein
|
| Buffer: |
25 mM Tris, 150 mM NaCl, pH: 8
|
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2020 Jan 25
|
SAXS reveals highly flexible interdomain linkers of tandem acyl carrier protein–thioesterase domains from a fungal nonreducing polyketide synthase
FEBS Letters (2020)
Bunnak W, Winter A, Lazarus C, Crump M, Race P, Wattana‐Amorn P
|
| RgGuinier |
3.6 |
nm |
| Dmax |
14.4 |
nm |
| VolumePorod |
75 |
nm3 |
|
|
|
|
|
|
|
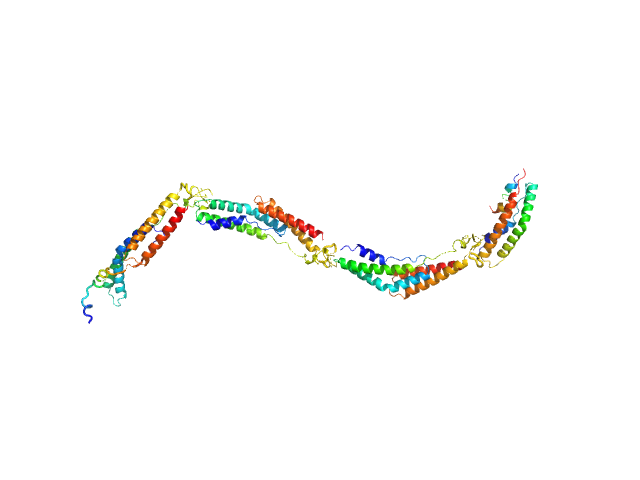
| Sample: |
Menisporopsin doublet Acyl Carrier Protein-Thioesterase monomer, 58 kDa Menisporopsis theobromae BCC … protein
|
| Buffer: |
25 mM Tris, 150 mM NaCl, pH: 8
|
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2020 Jan 25
|
SAXS reveals highly flexible interdomain linkers of tandem acyl carrier protein–thioesterase domains from a fungal nonreducing polyketide synthase
FEBS Letters (2020)
Bunnak W, Winter A, Lazarus C, Crump M, Race P, Wattana‐Amorn P
|
| RgGuinier |
4.2 |
nm |
| Dmax |
17.3 |
nm |
| VolumePorod |
84 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Extracellular matrix binding protein F-repeats , 70 kDa Staphylococcus epidermidis protein
|
| Buffer: |
50 mM MES, 150 mM NaCl, pH: 6
|
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 Dec 15
|
A Giant Extracellular Matrix Binding Protein of Staphylococcus epidermidis Binds Surface-Immobilized Fibronectin via a Novel Mechanism.
mBio 11(5) (2020)
Büttner H, Perbandt M, Kohler T, Kikhney A, Wolters M, Christner M, Heise M, Wilde J, Weißelberg S, Both A, Betzel C, Hammerschmidt S, Svergun D, Aepfelbacher M, Rohde H
|
| RgGuinier |
7.0 |
nm |
| Dmax |
28.0 |
nm |
|
|
|
|
|
|

|
| Sample: |
Extracellular matrix binding protein FG-repeats , 85 kDa Staphylococcus epidermidis protein
|
| Buffer: |
PBS, 136.5 mM NaCl, 2.65 mM KCl, 8.3 mM Na2HPO4, 2.65 mM KH2PO4, pH: 7.4
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Jun 14
|
A Giant Extracellular Matrix Binding Protein of Staphylococcus epidermidis Binds Surface-Immobilized Fibronectin via a Novel Mechanism.
mBio 11(5) (2020)
Büttner H, Perbandt M, Kohler T, Kikhney A, Wolters M, Christner M, Heise M, Wilde J, Weißelberg S, Both A, Betzel C, Hammerschmidt S, Svergun D, Aepfelbacher M, Rohde H
|
| RgGuinier |
10.7 |
nm |
| Dmax |
40.0 |
nm |
|
|
|
|
|
|
|
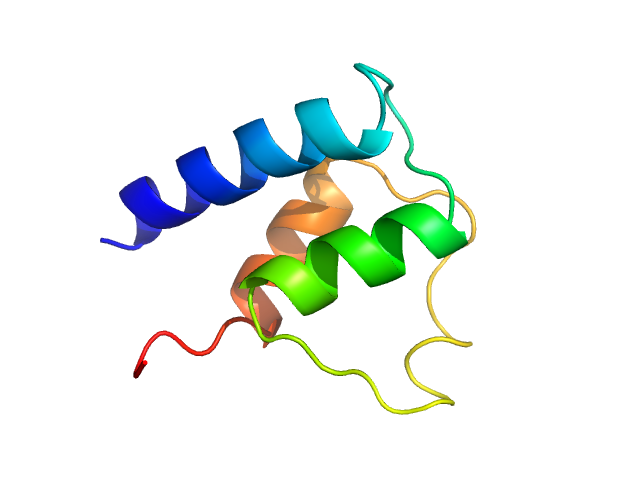
| Sample: |
ACT domain of Rel protein (Bifunctional (p)ppGpp synthase/hydrolase RelA) dimer, 20 kDa Mycobacterium tuberculosis protein
|
| Buffer: |
50 mM Tris-HCl, 350 mM NaCl, 5% glycerol, 1 mM DTT, pH: 8.5
|
| Experiment: |
SAXS
data collected at Bruker Nanostar, Nanyang Technological University on 2018 Jun 7
|
Atomic structure of, and valine binding to the regulatory ACT domain of the Mycobacterium tuberculosis Rel protein.
FEBS J (2020)
Shin J, Singal B, Manimekalai MSS, Chen MW, Ragunathan P, Grüber G
|
| RgGuinier |
1.9 |
nm |
| Dmax |
6.1 |
nm |
| VolumePorod |
29 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Myosin essential light chain monomer, 9 kDa Plasmodium falciparum protein
|
| Buffer: |
20 mM HEPES pH 7.5, 150 mM NaCl, 0.5 mM TCEP, pH: 7.5
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Oct 25
|
Structural role of essential light chains in the apicomplexan glideosome.
Commun Biol 3(1):568 (2020)
Pazicky S, Dhamotharan K, Kaszuba K, Mertens HDT, Gilberger T, Svergun D, Kosinski J, Weininger U, Löw C
|
| RgGuinier |
1.4 |
nm |
| Dmax |
4.3 |
nm |
| VolumePorod |
12 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Myosin essential light chain monomer, 16 kDa Plasmodium falciparum protein
|
| Buffer: |
20 mM HEPES pH 7.5, 150 mM NaCl, 0.5 mM TCEP, pH: 7.5
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Jun 30
|
Structural role of essential light chains in the apicomplexan glideosome.
Commun Biol 3(1):568 (2020)
Pazicky S, Dhamotharan K, Kaszuba K, Mertens HDT, Gilberger T, Svergun D, Kosinski J, Weininger U, Löw C
|
| RgGuinier |
2.7 |
nm |
| Dmax |
9.5 |
nm |
| VolumePorod |
23 |
nm3 |
|
|
|
|
|
|
|
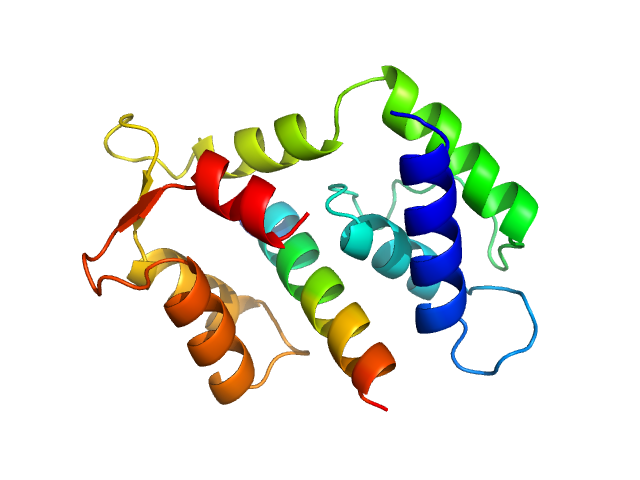
| Sample: |
Myosin essential light chain 2 monomer, 15 kDa Toxoplasma gondii protein
|
| Buffer: |
20 mM HEPES pH 7.5, 150 mM NaCl, 0.5 mM TCEP, pH: 7.5
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Apr 8
|
Structural role of essential light chains in the apicomplexan glideosome.
Commun Biol 3(1):568 (2020)
Pazicky S, Dhamotharan K, Kaszuba K, Mertens HDT, Gilberger T, Svergun D, Kosinski J, Weininger U, Löw C
|
| RgGuinier |
2.1 |
nm |
| Dmax |
6.7 |
nm |
| VolumePorod |
27 |
nm3 |
|
|
|
|
|
|
|
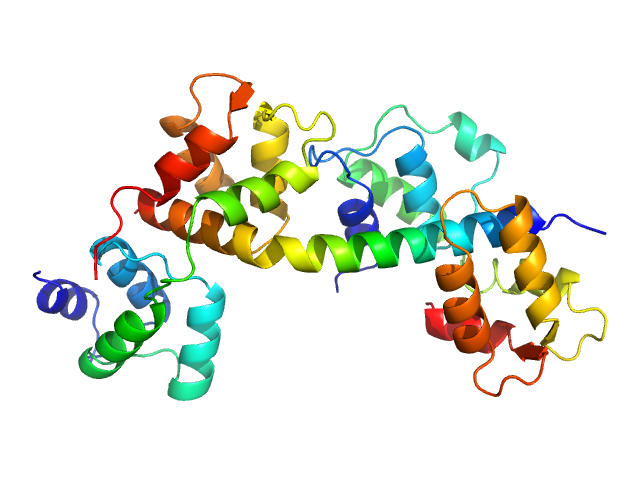
| Sample: |
Myosin essential light chain 2 monomer, 15 kDa Toxoplasma gondii protein
Myosin A monomer, 3 kDa Toxoplasma gondii protein
|
| Buffer: |
20 mM HEPES pH 7.5, 150 mM NaCl, 0.5 mM TCEP, pH: 7.5
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Apr 8
|
Structural role of essential light chains in the apicomplexan glideosome.
Commun Biol 3(1):568 (2020)
Pazicky S, Dhamotharan K, Kaszuba K, Mertens HDT, Gilberger T, Svergun D, Kosinski J, Weininger U, Löw C
|
| RgGuinier |
1.7 |
nm |
| Dmax |
5.5 |
nm |
| VolumePorod |
28 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Toxoplasma gondii essential light chain 1 monomer, 15 kDa Toxoplasma gondii protein
Myosin A monomer, 5 kDa Toxoplasma gondii protein
Toxoplasma gondii myosin light chain, full length monomer, 24 kDa Toxoplasma gondii protein
|
| Buffer: |
20 mM HEPES pH 7.5, 150 mM NaCl, 0.5 mM TCEP, pH: 7.5
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Sep 30
|
Structural role of essential light chains in the apicomplexan glideosome.
Commun Biol 3(1):568 (2020)
Pazicky S, Dhamotharan K, Kaszuba K, Mertens HDT, Gilberger T, Svergun D, Kosinski J, Weininger U, Löw C
|
| RgGuinier |
3.2 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
69 |
nm3 |
|
|