|
|
|
|
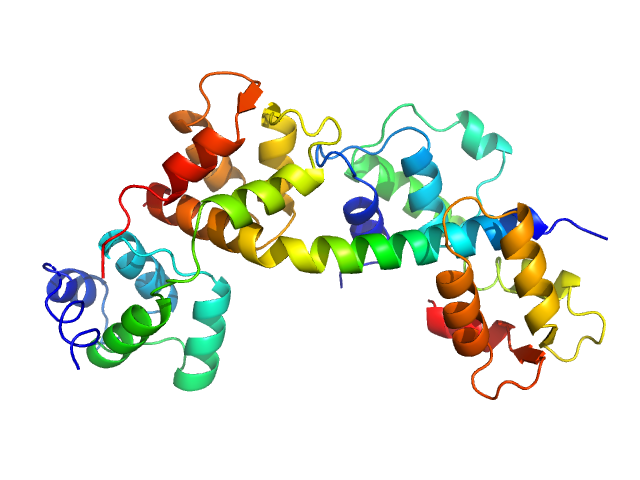
|
| Sample: |
Myosin light chain TgMLC1, residues 66-210 monomer, 17 kDa Toxoplasma gondii protein
Toxoplasma gondii essential light chain 1 monomer, 15 kDa Toxoplasma gondii protein
Myosin A monomer, 5 kDa Toxoplasma gondii protein
|
| Buffer: |
20 mM HEPES pH 7.5, 150 mM NaCl, 0.5 mM TCEP, pH: 7.5
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Sep 30
|
Structural role of essential light chains in the apicomplexan glideosome.
Commun Biol 3(1):568 (2020)
Pazicky S, Dhamotharan K, Kaszuba K, Mertens HDT, Gilberger T, Svergun D, Kosinski J, Weininger U, Löw C
|
| RgGuinier |
2.7 |
nm |
| Dmax |
9.5 |
nm |
| VolumePorod |
50 |
nm3 |
|
|
|
|
|
|
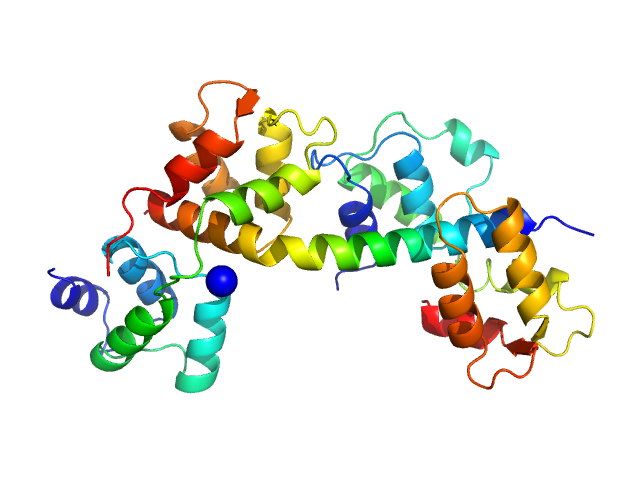
|
| Sample: |
Toxoplasma gondii essential light chain 1 monomer, 15 kDa Toxoplasma gondii protein
Myosin A monomer, 5 kDa Toxoplasma gondii protein
Toxoplasma gondii myosin light chain, residues 70-210 monomer, 17 kDa Toxoplasma gondii protein
|
| Buffer: |
20 mM HEPES pH 7.5, 150 mM NaCl, 0.5 mM TCEP, pH: 7.5
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Sep 30
|
Structural role of essential light chains in the apicomplexan glideosome.
Commun Biol 3(1):568 (2020)
Pazicky S, Dhamotharan K, Kaszuba K, Mertens HDT, Gilberger T, Svergun D, Kosinski J, Weininger U, Löw C
|
| RgGuinier |
2.5 |
nm |
| Dmax |
8.2 |
nm |
| VolumePorod |
47 |
nm3 |
|
|
|
|
|
|
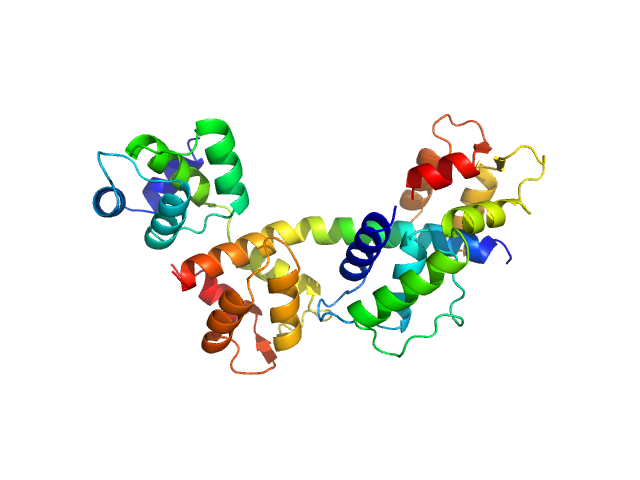
|
| Sample: |
Myosin essential light chain 2 monomer, 15 kDa Toxoplasma gondii protein
Myosin light chain TgMLC1, residues 66-210 monomer, 17 kDa Toxoplasma gondii protein
Myosin A monomer, 5 kDa Toxoplasma gondii protein
|
| Buffer: |
20 mM HEPES pH 7.5, 150 mM NaCl, 0.5 mM TCEP, pH: 7.5
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Sep 30
|
Structural role of essential light chains in the apicomplexan glideosome.
Commun Biol 3(1):568 (2020)
Pazicky S, Dhamotharan K, Kaszuba K, Mertens HDT, Gilberger T, Svergun D, Kosinski J, Weininger U, Löw C
|
| RgGuinier |
2.7 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
50 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Myosin essential light chain monomer, 16 kDa Plasmodium falciparum protein
Plasmodium falciparum myosin A monomer, 5 kDa Plasmodium falciparum protein
Myosin A tail domain interacting protein monomer, 17 kDa Plasmodium falciparum protein
|
| Buffer: |
20 mM HEPES pH 7.5, 150 mM NaCl, 0.5 mM TCEP, pH: 7.5
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Sep 30
|
Structural role of essential light chains in the apicomplexan glideosome.
Commun Biol 3(1):568 (2020)
Pazicky S, Dhamotharan K, Kaszuba K, Mertens HDT, Gilberger T, Svergun D, Kosinski J, Weininger U, Löw C
|
| RgGuinier |
2.7 |
nm |
| Dmax |
10.7 |
nm |
| VolumePorod |
51 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Ataxin-3 (polyglutamine protein ataxin-3 (Q13)) monomer, 41 kDa Homo sapiens protein
|
| Buffer: |
20 mM sodium phosphate buffer, 2 mM TCEP, 5% glycerol, pH: 6.5
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 May 24
|
Capturing the Conformational Ensemble of the Mixed Folded Polyglutamine Protein Ataxin-3.
Structure (2020)
Sicorello A, Różycki B, Konarev PV, Svergun DI, Pastore A
|
| RgGuinier |
4.0 |
nm |
| Dmax |
14.1 |
nm |
| VolumePorod |
79 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Ataxin-3 (polyglutamine protein ataxin-3 (Q54)) monomer, 46 kDa protein
|
| Buffer: |
20 mM sodium phosphate buffer, 2 mM TCEP, 5% glycerol, pH: 6.5
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 May 24
|
Capturing the Conformational Ensemble of the Mixed Folded Polyglutamine Protein Ataxin-3.
Structure (2020)
Sicorello A, Różycki B, Konarev PV, Svergun DI, Pastore A
|
| RgGuinier |
4.2 |
nm |
| Dmax |
15.5 |
nm |
| VolumePorod |
89 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Talin-1 (Δ139-168), human monomer, 48 kDa Homo sapiens protein
|
| Buffer: |
50 mM sodium phosphate, 150mM NaCl, pH: 7.2
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2015 Sep 24
|
The F1 loop of the talin head domain acts as a gatekeeper in integrin activation and clustering.
J Cell Sci 133(19) (2020)
Kukkurainen S, Azizi L, Zhang P, Jacquier MC, Baikoghli M, von Essen M, Tuukkanen A, Laitaoja M, Liu X, Rahikainen R, Orłowski A, Jänis J, Määttä JAE, Varjosalo M, Vattulainen I, Róg T, Svergun D, Che...
|
| RgGuinier |
3.3 |
nm |
| Dmax |
13.3 |
nm |
| VolumePorod |
77 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Talin-1, human monomer, 51 kDa Homo sapiens protein
|
| Buffer: |
50 mM sodium phosphate, 150 mM NaCl, pH: 7.2
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2015 Sep 24
|
The F1 loop of the talin head domain acts as a gatekeeper in integrin activation and clustering.
J Cell Sci 133(19) (2020)
Kukkurainen S, Azizi L, Zhang P, Jacquier MC, Baikoghli M, von Essen M, Tuukkanen A, Laitaoja M, Liu X, Rahikainen R, Orłowski A, Jänis J, Määttä JAE, Varjosalo M, Vattulainen I, Róg T, Svergun D, Che...
|
| RgGuinier |
3.4 |
nm |
| Dmax |
11.5 |
nm |
| VolumePorod |
94 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Talin-1 (Δ134-170/GAG insert), human monomer, 47 kDa Homo sapiens protein
|
| Buffer: |
50 mM sodium phosphate, 150mM NaCl, pH: 7.2
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2015 Sep 24
|
The F1 loop of the talin head domain acts as a gatekeeper in integrin activation and clustering.
J Cell Sci 133(19) (2020)
Kukkurainen S, Azizi L, Zhang P, Jacquier MC, Baikoghli M, von Essen M, Tuukkanen A, Laitaoja M, Liu X, Rahikainen R, Orłowski A, Jänis J, Määttä JAE, Varjosalo M, Vattulainen I, Róg T, Svergun D, Che...
|
| RgGuinier |
3.0 |
nm |
| Dmax |
10.2 |
nm |
| VolumePorod |
71 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Salt stress-induced protein dimer, 37 kDa Oryza sativa Indica … protein
|
| Buffer: |
50 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, pH: 7.4
|
| Experiment: |
SAXS
data collected at Anton Paar SAXSpace, CSIR - Institute of Microbial Technology (IMTech) on 2017 Mar 6
|
Structural insights into rice SalTol QTL located SALT protein.
Sci Rep 10(1):16589 (2020)
Kaur N, Sagar A, Sharma P, Ashish, Pati PK
|
| RgGuinier |
2.5 |
nm |
| Dmax |
6.7 |
nm |
| VolumePorod |
56 |
nm3 |
|
|