|
|
|
|

|
| Sample: |
Meiotic localizer of BRCA2 dimer, 26 kDa Mus musculus protein
|
| Buffer: |
20 mM Tris pH 8.0, 150 mM KCl, pH: 8
|
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Oct 7
|
The BRCA2-MEILB2-BRME1 complex governs meiotic recombination and impairs the mitotic BRCA2-RAD51 function in cancer cells
Nature Communications 11(1) (2020)
Zhang J, Gurusaran M, Fujiwara Y, Zhang K, Echbarthi M, Vorontsov E, Guo R, Pendlebury D, Alam I, Livera G, Emmanuelle M, Wang P, Nandakumar J, Davies O, Shibuya H
|
| RgGuinier |
4.6 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
88 |
nm3 |
|
|
|
|
|
|

|
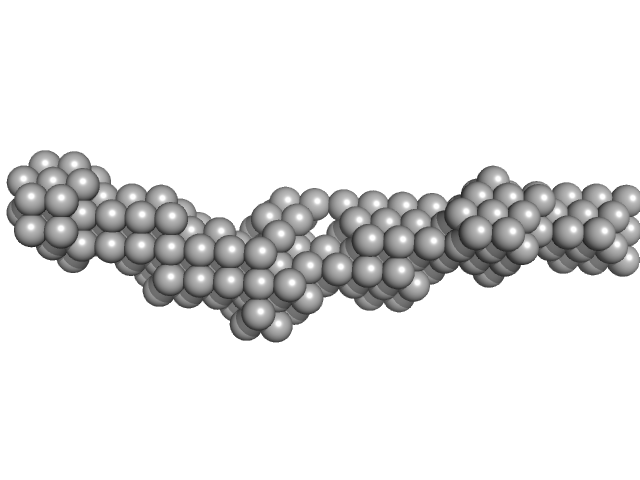
| Sample: |
Meiotic localizer of BRCA2 octamer, 102 kDa Mus musculus protein
|
| Buffer: |
20 mM Tris pH 8.0, 150 mM KCl, pH: 8
|
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Oct 7
|
The BRCA2-MEILB2-BRME1 complex governs meiotic recombination and impairs the mitotic BRCA2-RAD51 function in cancer cells
Nature Communications 11(1) (2020)
Zhang J, Gurusaran M, Fujiwara Y, Zhang K, Echbarthi M, Vorontsov E, Guo R, Pendlebury D, Alam I, Livera G, Emmanuelle M, Wang P, Nandakumar J, Davies O, Shibuya H
|
| RgGuinier |
5.7 |
nm |
| Dmax |
20.0 |
nm |
| VolumePorod |
368 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Meiotic localizer of BRCA2 monomer, 9 kDa Mus musculus protein
|
| Buffer: |
20 mM Tris pH 8.0, 150 mM KCl, pH: 8
|
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Oct 7
|
The BRCA2-MEILB2-BRME1 complex governs meiotic recombination and impairs the mitotic BRCA2-RAD51 function in cancer cells
Nature Communications 11(1) (2020)
Zhang J, Gurusaran M, Fujiwara Y, Zhang K, Echbarthi M, Vorontsov E, Guo R, Pendlebury D, Alam I, Livera G, Emmanuelle M, Wang P, Nandakumar J, Davies O, Shibuya H
|
| RgGuinier |
3.0 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
43 |
nm3 |
|
|
|
|
|
|
|
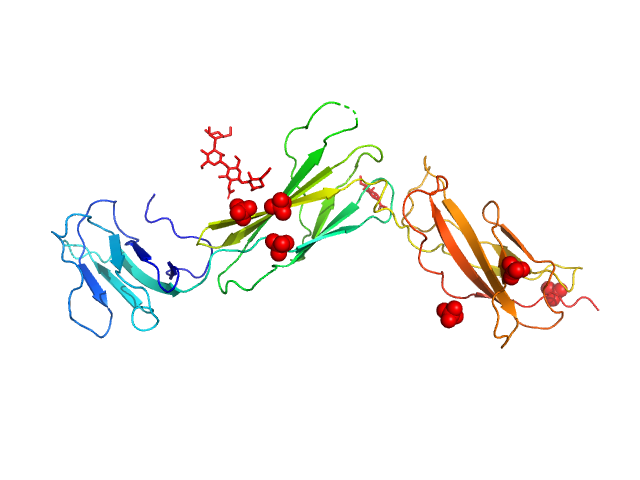
| Sample: |
Interleukin-11 receptor subunit alpha monomer, 32 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 0.2% sodium azide, pH: 8.5
|
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2019 Jun 8
|
The structure of the extracellular domains of human interleukin 11 α-receptor reveals mechanisms of cytokine engagement
Journal of Biological Chemistry :jbc.RA119.012351 (2020)
Metcalfe R, Aizel K, Zlatic C, Nguyen P, Morton C, Lio D, Cheng H, Dobson R, Parker M, Gooley P, Putoczki T, Griffin M
|
| RgGuinier |
3.0 |
nm |
| Dmax |
9.5 |
nm |
| VolumePorod |
41 |
nm3 |
|
|
|
|
|
|
|
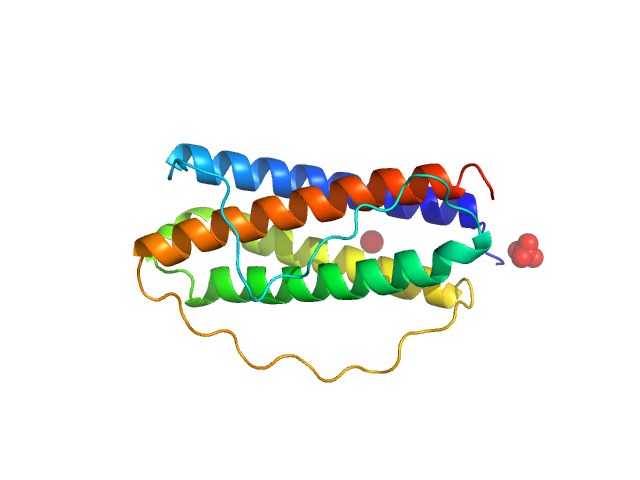
| Sample: |
Interleukin 11 monomer, 18 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 0.2% sodium azide, pH: 8.5
|
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2019 Jun 8
|
The structure of the extracellular domains of human interleukin 11 α-receptor reveals mechanisms of cytokine engagement
Journal of Biological Chemistry :jbc.RA119.012351 (2020)
Metcalfe R, Aizel K, Zlatic C, Nguyen P, Morton C, Lio D, Cheng H, Dobson R, Parker M, Gooley P, Putoczki T, Griffin M
|
| RgGuinier |
1.7 |
nm |
| Dmax |
5.4 |
nm |
| VolumePorod |
22 |
nm3 |
|
|
|
|
|
|
|
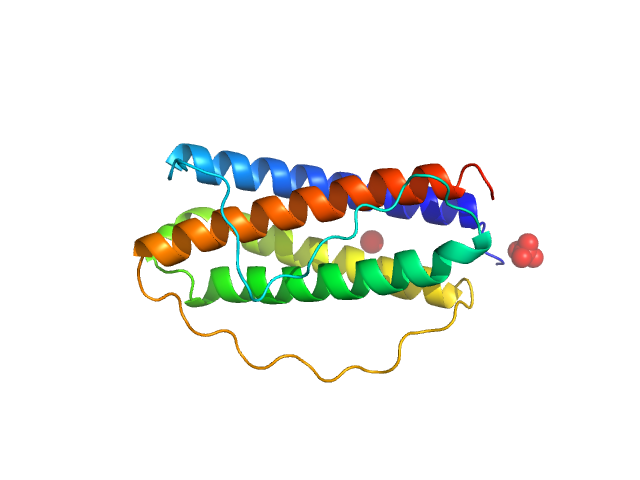
| Sample: |
Interleukin 11 monomer, 19 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 0.2% sodium azide, pH: 8.5
|
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2019 Jun 8
|
The structure of the extracellular domains of human interleukin 11 α-receptor reveals mechanisms of cytokine engagement
Journal of Biological Chemistry :jbc.RA119.012351 (2020)
Metcalfe R, Aizel K, Zlatic C, Nguyen P, Morton C, Lio D, Cheng H, Dobson R, Parker M, Gooley P, Putoczki T, Griffin M
|
| RgGuinier |
1.9 |
nm |
| Dmax |
6.1 |
nm |
| VolumePorod |
28 |
nm3 |
|
|
|
|
|
|
|
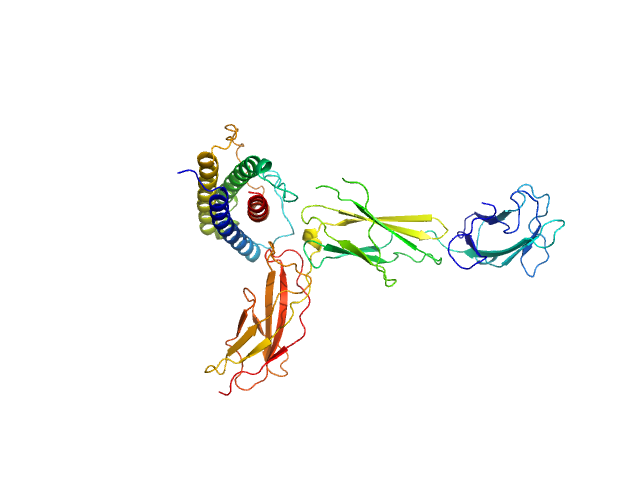
| Sample: |
Interleukin-11 receptor subunit alpha monomer, 32 kDa Homo sapiens protein
Interleukin 11 monomer, 18 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 0.2% sodium azide, pH: 8.5
|
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2019 Jun 8
|
The structure of the extracellular domains of human interleukin 11 α-receptor reveals mechanisms of cytokine engagement
Journal of Biological Chemistry :jbc.RA119.012351 (2020)
Metcalfe R, Aizel K, Zlatic C, Nguyen P, Morton C, Lio D, Cheng H, Dobson R, Parker M, Gooley P, Putoczki T, Griffin M
|
| RgGuinier |
3.3 |
nm |
| Dmax |
10.2 |
nm |
| VolumePorod |
84 |
nm3 |
|
|
|
|
|
|
|
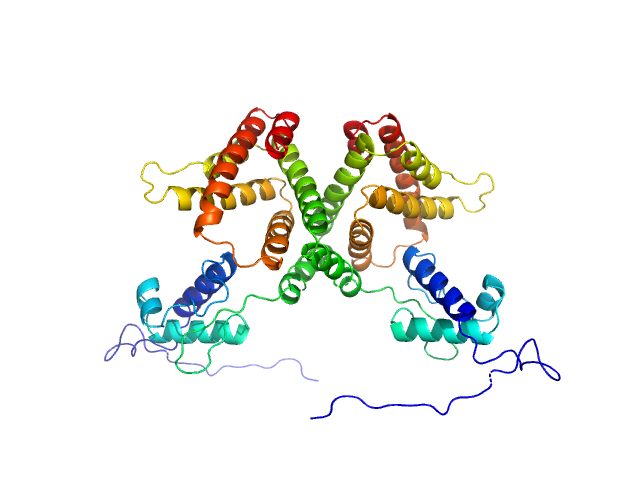
| Sample: |
HTH-type transcriptional repressor NanR dimer, 59 kDa Escherichia coli protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 0.1 % (w/v) sodium azide, pH: 8
|
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2019 Apr 20
|
Mechanism of NanR gene repression and allosteric induction of bacterial sialic acid metabolism
(2020)
Horne C, Venugopal H, Panjikar S, Henrickson A, Brookes E, North R, Murphy J, Friemann R, Griffin M, Ramm G, Demeler B, Dobson R
|
| RgGuinier |
3.2 |
nm |
| Dmax |
10.5 |
nm |
| VolumePorod |
110 |
nm3 |
|
|
|
|
|
|
|
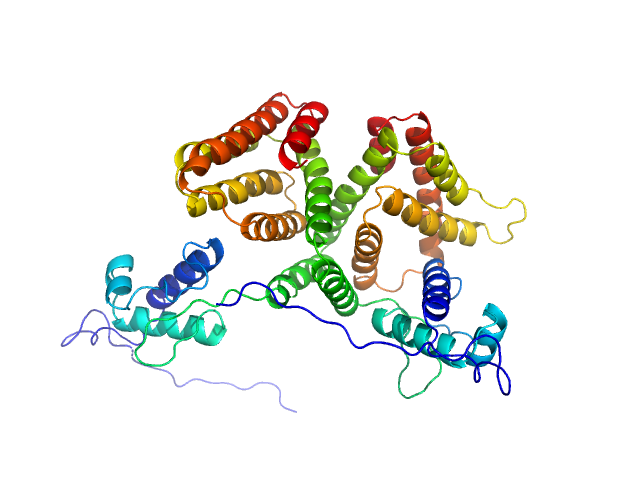
| Sample: |
HTH-type transcriptional repressor NanR dimer, 59 kDa Escherichia coli protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 20 mM Neu5Ac and 0.1 % (w/v) sodium azide, pH: 8
|
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2019 Apr 20
|
Mechanism of NanR gene repression and allosteric induction of bacterial sialic acid metabolism
(2020)
Horne C, Venugopal H, Panjikar S, Henrickson A, Brookes E, North R, Murphy J, Friemann R, Griffin M, Ramm G, Demeler B, Dobson R
|
| RgGuinier |
3.1 |
nm |
| Dmax |
10.1 |
nm |
| VolumePorod |
105 |
nm3 |
|
|
|
|
|
|
|
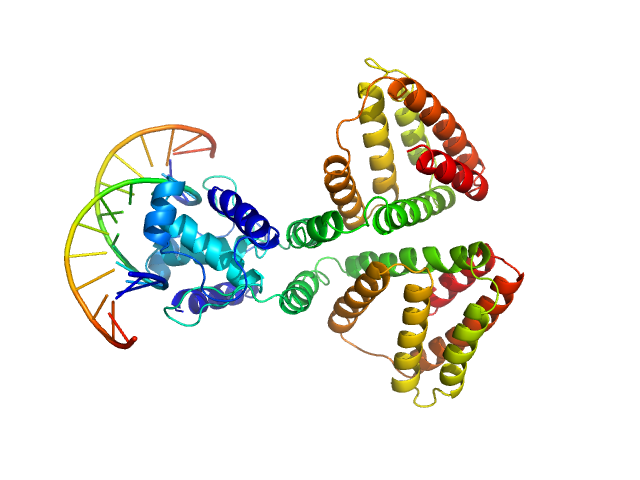
| Sample: |
HTH-type transcriptional repressor NanR dimer, 59 kDa Escherichia coli protein
(GGTATA)2 repeat DNA monomer, 11 kDa DNA
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 0.1 % (w/v) sodium azide, pH: 8
|
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2019 Apr 20
|
Mechanism of NanR gene repression and allosteric induction of bacterial sialic acid metabolism
(2020)
Horne C, Venugopal H, Panjikar S, Henrickson A, Brookes E, North R, Murphy J, Friemann R, Griffin M, Ramm G, Demeler B, Dobson R
|
| RgGuinier |
3.3 |
nm |
| Dmax |
9.8 |
nm |
| VolumePorod |
108 |
nm3 |
|
|