|
|
|
|

|
| Sample: |
Antiparallel Hairpin monomer, 21 kDa synthetic construct DNA
|
| Buffer: |
6 mM Na2HPO4, 2 mM NaH2PO4, 1 mM Na2EDTA, 185 mM KCl, pH: 7.2
|
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2019 Nov 3
|
The hTERT core promoter forms three parallel G-quadruplexes.
Nucleic Acids Res (2020)
Monsen RC, DeLeeuw L, Dean WL, Gray RD, Sabo TM, Chakravarthy S, Chaires JB, Trent JO
|
| RgGuinier |
2.5 |
nm |
| Dmax |
8.1 |
nm |
| VolumePorod |
28 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Optimized Parallel monomer, 18 kDa synthetic construct DNA
|
| Buffer: |
6 mM Na2HPO4, 2 mM NaH2PO4, 1 mM Na2EDTA, 185 mM KCl, pH: 7.2
|
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2019 Nov 3
|
The hTERT core promoter forms three parallel G-quadruplexes.
Nucleic Acids Res (2020)
Monsen RC, DeLeeuw L, Dean WL, Gray RD, Sabo TM, Chakravarthy S, Chaires JB, Trent JO
|
| RgGuinier |
1.7 |
nm |
| Dmax |
5.0 |
nm |
| VolumePorod |
17 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
hTERT core promoter PQS2-PQS3 region monomer, 14 kDa synthetic construct DNA
|
| Buffer: |
6 mM Na2HPO4, 2 mM NaH2PO4, 1 mM Na2EDTA, 185 mM KCl, pH: 7.2
|
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2019 Nov 3
|
The hTERT core promoter forms three parallel G-quadruplexes.
Nucleic Acids Res (2020)
Monsen RC, DeLeeuw L, Dean WL, Gray RD, Sabo TM, Chakravarthy S, Chaires JB, Trent JO
|
| RgGuinier |
1.7 |
nm |
| Dmax |
5.4 |
nm |
| VolumePorod |
18 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Antiparallel Hairpin PQS2-PQS3 segment monomer, 14 kDa synthetic construct DNA
|
| Buffer: |
6 mM Na2HPO4, 2 mM NaH2PO4, 1 mM Na2EDTA, 185 mM KCl, pH: 7.2
|
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2019 Nov 3
|
The hTERT core promoter forms three parallel G-quadruplexes.
Nucleic Acids Res (2020)
Monsen RC, DeLeeuw L, Dean WL, Gray RD, Sabo TM, Chakravarthy S, Chaires JB, Trent JO
|
| RgGuinier |
2.0 |
nm |
| Dmax |
6.4 |
nm |
| VolumePorod |
18 |
nm3 |
|
|
|
|
|
|
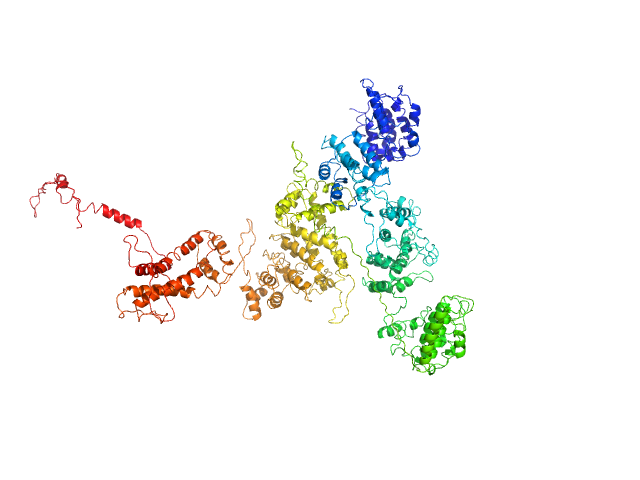
|
| Sample: |
Macrophage mannose receptor 1 dimer, 315 kDa Mouse myeloma cell … protein
|
| Buffer: |
50mM Hepes, 100mM NaCl, 1mM DTT, pH: 7
|
| Experiment: |
SAXS
data collected at 12-ID-B SAXS/WAXS, Advanced Photon Source (APS), Argonne National Laboratory on 2016 Apr 15
|
Mannose receptor (CD206) activation in tumor-associated macrophages enhances adaptive and innate antitumor immune responses.
Sci Transl Med 12(530) (2020)
Jaynes JM, Sable R, Ronzetti M, Bautista W, Knotts Z, Abisoye-Ogunniyan A, Li D, Calvo R, Dashnyam M, Singh A, Guerin T, White J, Ravichandran S, Kumar P, Talsania K, Chen V, Ghebremedhin A, Karanam B...
|
| RgGuinier |
7.9 |
nm |
| Dmax |
30.1 |
nm |
| VolumePorod |
584 |
nm3 |
|
|
|
|
|
|
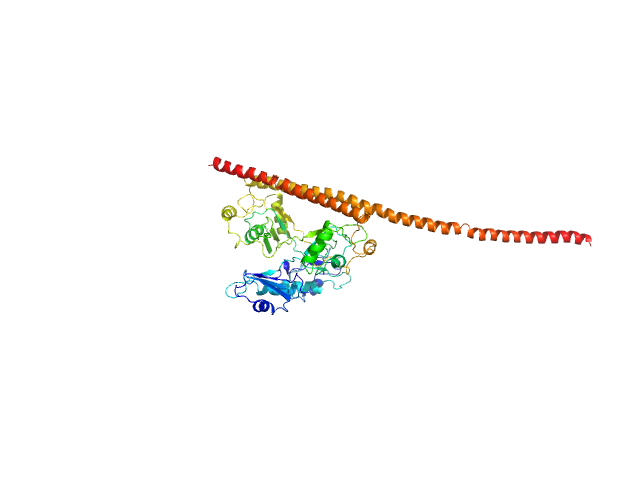
|
| Sample: |
Non-POU domain-containing octamer-binding protein monomer, 30 kDa Homo sapiens protein
Splicing factor, proline- and glutamine-rich monomer, 38 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 250 mM NaCl, pH: 7.5
|
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2018 Apr 5
|
Structural basis of the zinc-induced cytoplasmic aggregation of the RNA-binding protein SFPQ
Nucleic Acids Research 48(6):3356-3365 (2020)
Huang J, Ringuet M, Whitten A, Caria S, Lim Y, Badhan R, Anggono V, Lee M
|
| RgGuinier |
4.0 |
nm |
| Dmax |
16.5 |
nm |
| VolumePorod |
90 |
nm3 |
|
|
|
|
|
|
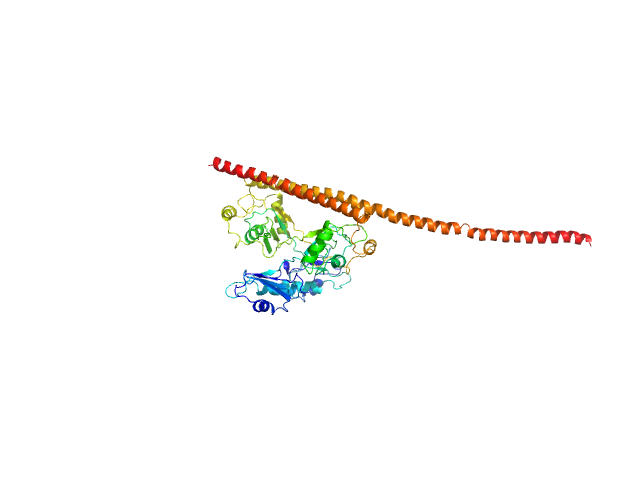
|
| Sample: |
Splicing factor, proline- and glutamine-rich monomer, 38 kDa Homo sapiens protein
Non-POU domain-containing octamer-binding protein monomer, 30 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 250 mM NaCl, pH: 7.5
|
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2018 Nov 8
|
Structural basis of the zinc-induced cytoplasmic aggregation of the RNA-binding protein SFPQ
Nucleic Acids Research 48(6):3356-3365 (2020)
Huang J, Ringuet M, Whitten A, Caria S, Lim Y, Badhan R, Anggono V, Lee M
|
| RgGuinier |
3.6 |
nm |
| Dmax |
16.5 |
nm |
| VolumePorod |
80 |
nm3 |
|
|
|
|
|
|
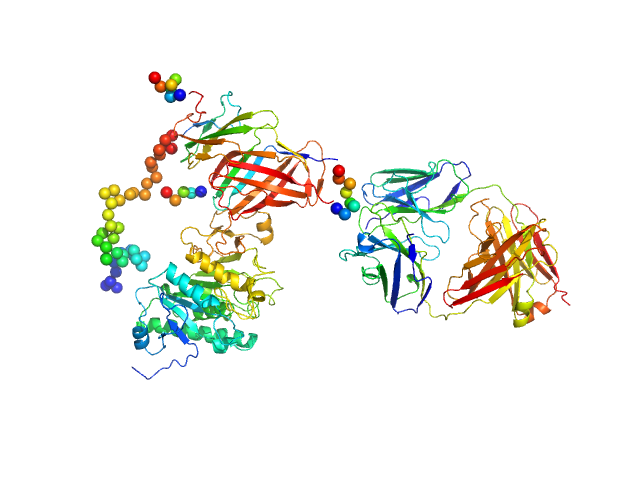
|
| Sample: |
Lipoprotein lipase monomer, 50 kDa Homo sapiens protein
Glycosylphosphatidylinositol-anchored high density lipoprotein-binding protein 1 monomer, 15 kDa Homo sapiens protein
Monoclonal Antibody Fragment 5D2 monomer, 47 kDa Mus musculus protein
|
| Buffer: |
10 mM Tris, 150 mM NaCl, 4 mM CaCL2, 10% (v/v) Glycerol, 0.05% 0.8mM CHAPS, ,0.05 % (v/v) NaN3, pH: 7.2
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Dec 14
|
Unfolding of monomeric lipoprotein lipase by ANGPTL4: Insight into the regulation of plasma triglyceride metabolism
Proceedings of the National Academy of Sciences :201920202 (2020)
Kristensen K, Leth-Espensen K, Mertens H, Birrane G, Meiyappan M, Olivecrona G, Jørgensen T, Young S, Ploug M
|
| RgGuinier |
4.9 |
nm |
| Dmax |
17.5 |
nm |
| VolumePorod |
187 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Latency associated peptide dimer, 58 kDa Homo sapiens protein
|
| Buffer: |
phosphate buffered saline 2% glycerol, pH: 7.4
|
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 Apr 20
|
Structural insights into conformational switching in latency-associated peptide between transforming growth factor β-1 bound and unbound states
IUCrJ 7(2) (2020)
Stachowski T, Snell M, Snell E
|
| RgGuinier |
3.5 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
129 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
lysozyme amyloid fibril , 14 kDa Gallus gallus protein
|
| Buffer: |
0.2 M glycine-HCl, 80 mM NaCl, pH: 2.2
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Sep 5
|
Effect of the concentration of protein and nanoparticles on the structure of biohybrid nanocomposites.
Biopolymers 111(2):e23342 (2020)
Majorošová J, Schroer MA, Tomašovičová N, Batková M, Hu PS, Kubovčíková M, Svergun DI, Kopčanský P
|
| RgGuinier |
30.2 |
nm |
| Dmax |
120.0 |
nm |
|
|