|
|
|
|
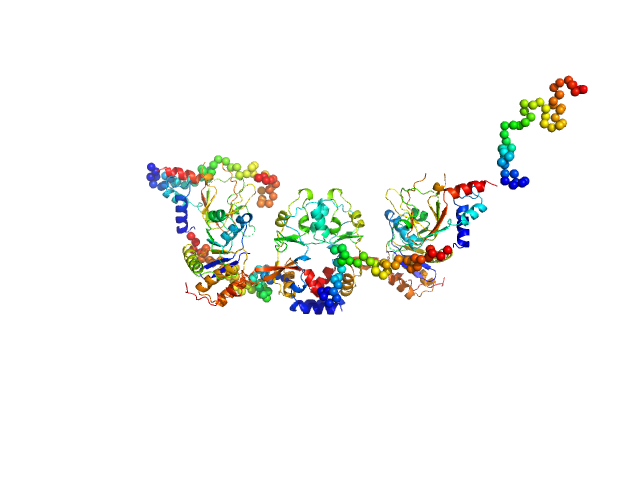
|
| Sample: |
CNNM4_BAT-cNMP-Ctail dimer, 96 kDa Homo sapiens protein
Protein tyrosine phosphatase type IVA 1 dimer, 40 kDa Mus musculus protein
|
| Buffer: |
HEPES buffer pH 7.4, 200 mM NaCl , 1mM DTT, pH: 7.4
|
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 Aug 2
|
Structural Insights into the Intracellular Region of the Human Magnesium Transport Mediator CNNM4.
Int J Mol Sci 20(24) (2019)
Giménez-Mascarell P, Oyenarte I, González-Recio I, Fernández-Rodríguez C, Corral-Rodríguez MÁ, Campos-Zarraga I, Simón J, Kostantin E, Hardy S, Díaz Quintana A, Zubillaga Lizeaga M, Merino N, Diercks ...
|
| RgGuinier |
4.7 |
nm |
| Dmax |
18.5 |
nm |
| VolumePorod |
226 |
nm3 |
|
|
|
|
|
|
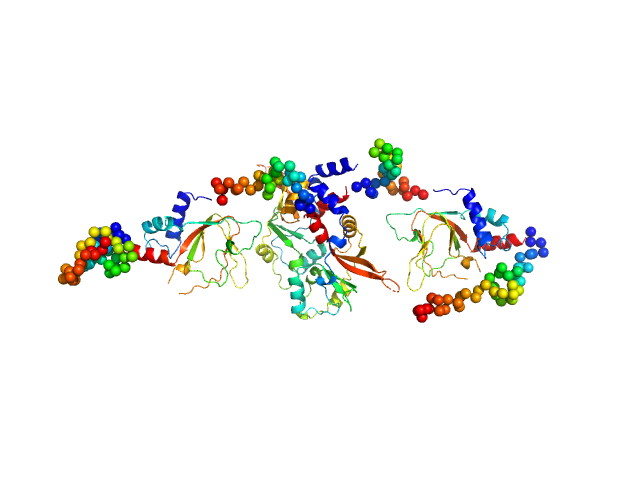
|
| Sample: |
CNNM4_BAT-cNMP-Ctail dimer, 96 kDa Homo sapiens protein
|
| Buffer: |
HEPES buffer pH 7.4, 200 mM NaCl , 1mM DTT, pH: 7.4
|
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 Aug 2
|
Structural Insights into the Intracellular Region of the Human Magnesium Transport Mediator CNNM4.
Int J Mol Sci 20(24) (2019)
Giménez-Mascarell P, Oyenarte I, González-Recio I, Fernández-Rodríguez C, Corral-Rodríguez MÁ, Campos-Zarraga I, Simón J, Kostantin E, Hardy S, Díaz Quintana A, Zubillaga Lizeaga M, Merino N, Diercks ...
|
| RgGuinier |
4.0 |
nm |
| Dmax |
16.6 |
nm |
| VolumePorod |
156 |
nm3 |
|
|
|
|
|
|
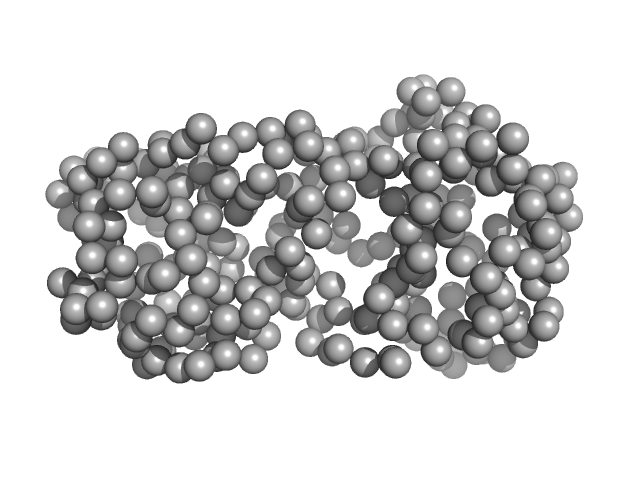
|
| Sample: |
CNNM4_cNMP dimer, 42 kDa Homo sapiens protein
|
| Buffer: |
HEPES buffer pH 7.4, 200 mM NaCl , 1mM DTT, pH: 7.4
|
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 Feb 7
|
Structural Insights into the Intracellular Region of the Human Magnesium Transport Mediator CNNM4.
Int J Mol Sci 20(24) (2019)
Giménez-Mascarell P, Oyenarte I, González-Recio I, Fernández-Rodríguez C, Corral-Rodríguez MÁ, Campos-Zarraga I, Simón J, Kostantin E, Hardy S, Díaz Quintana A, Zubillaga Lizeaga M, Merino N, Diercks ...
|
| RgGuinier |
2.6 |
nm |
| Dmax |
9.7 |
nm |
| VolumePorod |
102 |
nm3 |
|
|
|
|
|
|
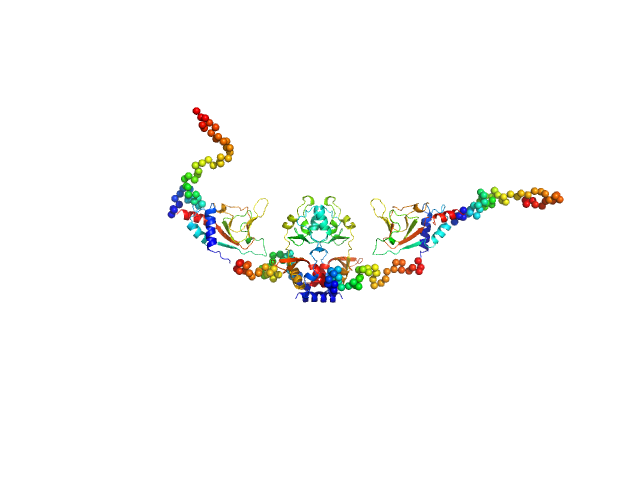
|
| Sample: |
CNNM4_BAT-cNMP-Ctail dimer, 96 kDa Homo sapiens protein
|
| Buffer: |
HEPES buffer pH 7.4, 200 mM NaCl , 1mM DTT, pH: 7.4
|
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 Sep 17
|
Structural Insights into the Intracellular Region of the Human Magnesium Transport Mediator CNNM4.
Int J Mol Sci 20(24) (2019)
Giménez-Mascarell P, Oyenarte I, González-Recio I, Fernández-Rodríguez C, Corral-Rodríguez MÁ, Campos-Zarraga I, Simón J, Kostantin E, Hardy S, Díaz Quintana A, Zubillaga Lizeaga M, Merino N, Diercks ...
|
| RgGuinier |
3.9 |
nm |
| Dmax |
14.3 |
nm |
| VolumePorod |
149 |
nm3 |
|
|
|
|
|
|
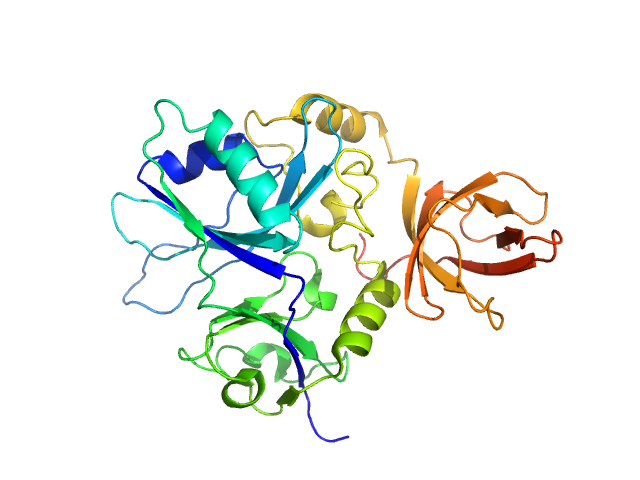
|
| Sample: |
Putative transferase CAF17, mitochondrial monomer, 33 kDa Homo sapiens protein
|
| Buffer: |
50 mM phosphate, 150 mM NaCl, 5 mM DTT, pH: 7
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Jul 10
|
Structural properties of [2Fe-2S] ISCA2-IBA57: a complex of the mitochondrial iron-sulfur cluster assembly machinery
Scientific Reports 9(1) (2019)
Nasta V, Da Vela S, Gourdoupis S, Ciofi-Baffoni S, Svergun D, Banci L
|
| RgGuinier |
2.2 |
nm |
| Dmax |
7.1 |
nm |
| VolumePorod |
50 |
nm3 |
|
|
|
|
|
|
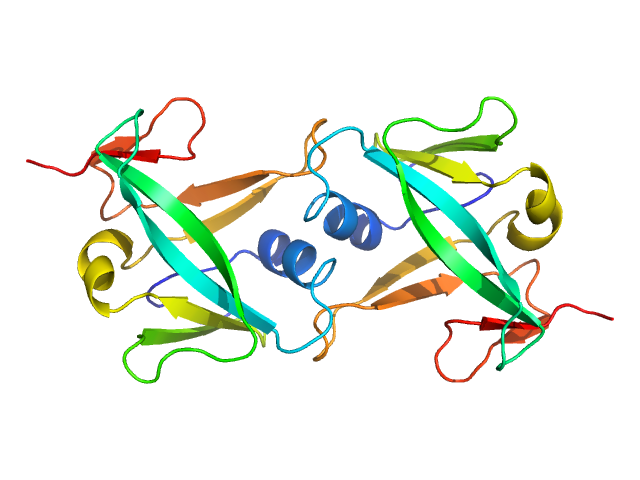
|
| Sample: |
Iron-sulfur cluster assembly 2 homolog, mitochondrial dimer, 24 kDa Homo sapiens protein
|
| Buffer: |
50 mM phosphate, 150 mM NaCl, 5 mM DTT, pH: 7
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Sep 16
|
Structural properties of [2Fe-2S] ISCA2-IBA57: a complex of the mitochondrial iron-sulfur cluster assembly machinery
Scientific Reports 9(1) (2019)
Nasta V, Da Vela S, Gourdoupis S, Ciofi-Baffoni S, Svergun D, Banci L
|
| RgGuinier |
2.0 |
nm |
| Dmax |
6.7 |
nm |
| VolumePorod |
31 |
nm3 |
|
|
|
|
|
|
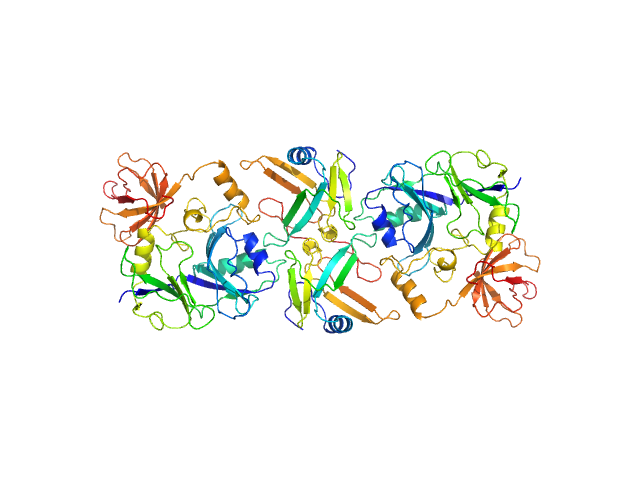
|
| Sample: |
Putative transferase CAF17, mitochondrial monomer, 33 kDa Homo sapiens protein
Iron-sulfur cluster assembly 2 homolog, mitochondrial dimer, 24 kDa Homo sapiens protein
|
| Buffer: |
50 mM phosphate, 150 mM NaCl, 5 mM DTT, pH: 7
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Nov 22
|
Structural properties of [2Fe-2S] ISCA2-IBA57: a complex of the mitochondrial iron-sulfur cluster assembly machinery
Scientific Reports 9(1) (2019)
Nasta V, Da Vela S, Gourdoupis S, Ciofi-Baffoni S, Svergun D, Banci L
|
| RgGuinier |
4.0 |
nm |
| Dmax |
16.4 |
nm |
| VolumePorod |
114 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Bruton's tyrosine kinase, kinase domain monomer, 32 kDa Homo sapiens protein
|
| Buffer: |
20mM Tris, 150mM NaCl, 1mM TCEP, 5% glycerol, pH: 7.5
|
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 May 11
|
Btk SH2-kinase interface is critical for allosteric kinase activation and its targeting inhibits B-cell neoplasms
Nature Communications 11(1) (2020)
Duarte D, Lamontanara A, La Sala G, Jeong S, Sohn Y, Panjkovich A, Georgeon S, Kükenshöner T, Marcaida M, Pojer F, De Vivo M, Svergun D, Kim H, Dal Peraro M, Hantschel O
|
| RgGuinier |
2.1 |
nm |
| Dmax |
6.8 |
nm |
| VolumePorod |
52 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Bruton's tyrosine kinase - Src homolgy domain 2 - kinase domain monomer, 46 kDa Homo sapiens protein
|
| Buffer: |
20mM Tris, 150mM NaCl, 1mM TCEP, 5% glycerol, pH: 7.5
|
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Nov 3
|
Btk SH2-kinase interface is critical for allosteric kinase activation and its targeting inhibits B-cell neoplasms
Nature Communications 11(1) (2020)
Duarte D, Lamontanara A, La Sala G, Jeong S, Sohn Y, Panjkovich A, Georgeon S, Kükenshöner T, Marcaida M, Pojer F, De Vivo M, Svergun D, Kim H, Dal Peraro M, Hantschel O
|
| RgGuinier |
2.8 |
nm |
| Dmax |
9.4 |
nm |
| VolumePorod |
64 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Bruton's tyrosine kinase - Src homology 3-2 kinase domain monomer, 52 kDa Homo sapiens protein
|
| Buffer: |
20mM Tris, 150mM NaCl, 1mM TCEP, 5% glycerol, pH: 7.5
|
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Nov 2
|
Btk SH2-kinase interface is critical for allosteric kinase activation and its targeting inhibits B-cell neoplasms
Nature Communications 11(1) (2020)
Duarte D, Lamontanara A, La Sala G, Jeong S, Sohn Y, Panjkovich A, Georgeon S, Kükenshöner T, Marcaida M, Pojer F, De Vivo M, Svergun D, Kim H, Dal Peraro M, Hantschel O
|
| RgGuinier |
2.6 |
nm |
| Dmax |
8.3 |
nm |
| VolumePorod |
72 |
nm3 |
|
|