|
|
|
|
|
| Sample: |
EKC/KEOPS complex subunit GON7 monomer, 13 kDa Homo sapiens protein
|
| Buffer: |
20 mM MES, 200 mM NaCl, 5 mM β-mercaptoethanol, pH: 6.5
|
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2017 Mar 26
|
Defects in t6A tRNA modification due to GON7 and YRDC mutations lead to Galloway-Mowat syndrome.
Nat Commun 10(1):3967 (2019)
Arrondel C, Missoury S, Snoek R, Patat J, Menara G, Collinet B, Liger D, Durand D, Gribouval O, Boyer O, Buscara L, Martin G, Machuca E, Nevo F, Lescop E, Braun DA, Boschat AC, Sanquer S, Guerrera IC,...
|
| RgGuinier |
3.1 |
nm |
| Dmax |
12.5 |
nm |
| VolumePorod |
46 |
nm3 |
|
|
|
|
|
|
|
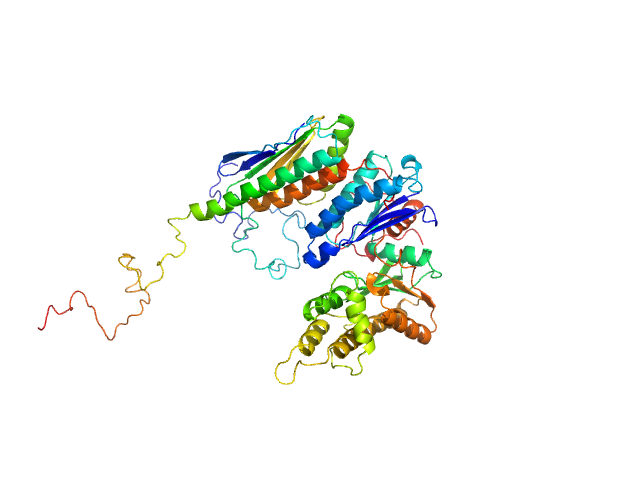
| Sample: |
EKC/KEOPS complex subunit GON7 monomer, 12 kDa Homo sapiens protein
EKC/KEOPS complex subunit LAGE3 monomer, 15 kDa Homo sapiens protein
Probable tRNA N6-adenosine threonylcarbamoyltransferase monomer, 36 kDa Homo sapiens protein
|
| Buffer: |
20 mM MES, 200 mM NaCl, 5 mM β-mercaptoethanol, pH: 6.5
|
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2017 May 12
|
Defects in t6A tRNA modification due to GON7 and YRDC mutations lead to Galloway-Mowat syndrome.
Nat Commun 10(1):3967 (2019)
Arrondel C, Missoury S, Snoek R, Patat J, Menara G, Collinet B, Liger D, Durand D, Gribouval O, Boyer O, Buscara L, Martin G, Machuca E, Nevo F, Lescop E, Braun DA, Boschat AC, Sanquer S, Guerrera IC,...
|
| RgGuinier |
3.1 |
nm |
| Dmax |
11.5 |
nm |
| VolumePorod |
91 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
EKC/KEOPS complex subunit GON7 dimer, 25 kDa Homo sapiens protein
EKC/KEOPS complex subunit LAGE3 dimer, 33 kDa Homo sapiens protein
|
| Buffer: |
20 mM MES, 200 mM NaCl, 5 mM β-mercaptoethanol, pH: 6.5
|
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2017 Mar 19
|
Defects in t6A tRNA modification due to GON7 and YRDC mutations lead to Galloway-Mowat syndrome.
Nat Commun 10(1):3967 (2019)
Arrondel C, Missoury S, Snoek R, Patat J, Menara G, Collinet B, Liger D, Durand D, Gribouval O, Boyer O, Buscara L, Martin G, Machuca E, Nevo F, Lescop E, Braun DA, Boschat AC, Sanquer S, Guerrera IC,...
|
| RgGuinier |
3.6 |
nm |
| Dmax |
15.5 |
nm |
| VolumePorod |
126 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Homeobox protein TGIF1 dimer, 23 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, pH: 7.4
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Oct 18
|
'TG' interacting factor
Ewelina Guca
|
| RgGuinier |
2.7 |
nm |
| Dmax |
9.2 |
nm |
| VolumePorod |
42 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Calredoxin, Redox protein from Chlamydomonas reinhardtii monomer, 40 kDa Chlamydomonas reinhardtii protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 1 mM DTT, 5 mM EGTA, pH: 8
|
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-1000, Structural Biology Laboratory, Graduate School of Medical Life Science, Yokohama City University on 2015 Nov 17
|
Calcium sensing via EF-hand 4 enables thioredoxin activity in the sensor-responder protein calredoxin in the green alga Chlamydomonas reinhardtii.
J Biol Chem (2019)
Charoenwattanasatien R, Zinzius K, Scholz M, Wicke S, Tanaka H, Brandenburg JS, Marchetti GM, Ikegami T, Matsumoto T, Oda T, Sato M, Hippler M, Kurisu G
|
| RgGuinier |
2.5 |
nm |
| Dmax |
8.7 |
nm |
| VolumePorod |
60 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Calredoxin, Redox protein from Chlamydomonas reinhardtii monomer, 40 kDa Chlamydomonas reinhardtii protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 1 mM DTT, 5 mM CaCl2, pH: 8
|
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-1000, Structural Biology Laboratory, Graduate School of Medical Life Science, Yokohama City University on 2015 Nov 17
|
Calcium sensing via EF-hand 4 enables thioredoxin activity in the sensor-responder protein calredoxin in the green alga Chlamydomonas reinhardtii.
J Biol Chem (2019)
Charoenwattanasatien R, Zinzius K, Scholz M, Wicke S, Tanaka H, Brandenburg JS, Marchetti GM, Ikegami T, Matsumoto T, Oda T, Sato M, Hippler M, Kurisu G
|
| RgGuinier |
3.1 |
nm |
| Dmax |
11.6 |
nm |
| VolumePorod |
68 |
nm3 |
|
|
|
|
|
|
|
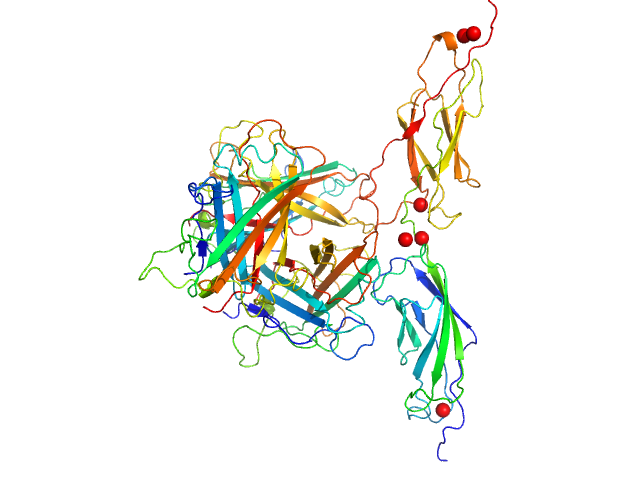
| Sample: |
Human adenovirus serotype 3 fibre knob trimer, 69 kDa Human adenovirus serotype … protein
Human desmoglein 2 extracellular domain 2 and 3 dimer, 55 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, 3 mM CaCl2, pH: 8
|
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Jan 26
|
Intermediate-resolution crystal structure of the human adenovirus B serotype 3 fibre knob in complex with the EC2-EC3 fragment of desmoglein 2.
Acta Crystallogr F Struct Biol Commun 75(Pt 12):750-757 (2019)
Vassal-Stermann E, Hutin S, Fender P, Burmeister WP
|
| RgGuinier |
3.4 |
nm |
| Dmax |
11.2 |
nm |
| VolumePorod |
188 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Cell wall synthesis protein Wag31 , 30 kDa Mycobacterium tuberculosis protein
|
| Buffer: |
50mM Tris pH7.5, 300mM NaCl, 10% Glycerol, 1mM EDTA (ethylene diamine tetra acetic acid), 5mM β-mercaptoethanol (BME), pH: 7.5
|
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Dec 15
|
Higher order assembling of the mycobacterial polar growth factor DivIVA/Wag31.
J Struct Biol :107429 (2019)
Choukate K, Gupta A, Basu B, Virk K, Ganguli M, Chaudhuri B
|
| RgGuinier |
19.8 |
nm |
| Dmax |
23.4 |
nm |
|
|
|
|
|
|
|
| Sample: |
Cell wall synthesis protein Wag31 , 30 kDa Mycobacterium tuberculosis protein
|
| Buffer: |
50mM Tris pH7.5, 300mM NaCl, 10% Glycerol, 1mM EDTA (ethylene diamine tetra acetic acid), 5mM β-mercaptoethanol (BME), pH: 7.5
|
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Dec 15
|
Higher order assembling of the mycobacterial polar growth factor DivIVA/Wag31.
J Struct Biol :107429 (2019)
Choukate K, Gupta A, Basu B, Virk K, Ganguli M, Chaudhuri B
|
| RgGuinier |
21.8 |
nm |
| Dmax |
13.6 |
nm |
|
|
|
|
|
|
|
| Sample: |
Cell wall synthesis protein Wag31 , 30 kDa Mycobacterium tuberculosis protein
|
| Buffer: |
50mM Tris pH7.5, 300mM NaCl, 10% Glycerol, 1mM EDTA (ethylene diamine tetra acetic acid), 5mM β-mercaptoethanol (BME), pH: 7.5
|
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2018 Jan 29
|
Higher order assembling of the mycobacterial polar growth factor DivIVA/Wag31.
J Struct Biol :107429 (2019)
Choukate K, Gupta A, Basu B, Virk K, Ganguli M, Chaudhuri B
|
|
|