|
|
|
|
|
| Sample: |
Adenylosuccinate lyase tetramer, 221 kDa Homo sapiens neanderthalensis protein
|
| Buffer: |
10 mM HEPES, 100 mM NaCl, 1 mM DTT, 1 mM AMP, pH: 7.5
|
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Jul 24
|
Molecular comparison of Neanderthal and Modern Human adenylosuccinate lyase.
Sci Rep 8(1):18008 (2018)
Van Laer B, Kapp U, Soler-Lopez M, Moczulska K, Pääbo S, Leonard G, Mueller-Dieckmann C
|
| RgGuinier |
3.7 |
nm |
| VolumePorod |
294 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Adenylosuccinate lyase tetramer, 221 kDa Homo sapiens neanderthalensis protein
|
| Buffer: |
10 mM HEPES, 100 mM NaCl, 1 mM DTT, 1 mM 5-aminoimidazole-4-carboxamide ribonucleotide (AICAR), pH: 7.5
|
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Jul 24
|
Molecular comparison of Neanderthal and Modern Human adenylosuccinate lyase.
Sci Rep 8(1):18008 (2018)
Van Laer B, Kapp U, Soler-Lopez M, Moczulska K, Pääbo S, Leonard G, Mueller-Dieckmann C
|
| RgGuinier |
3.7 |
nm |
| VolumePorod |
292 |
nm3 |
|
|
|
|
|
|
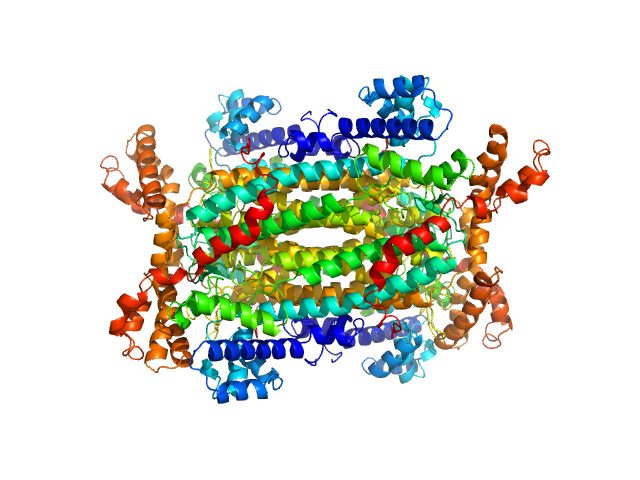
|
| Sample: |
Adenylosuccinate lyase tetramer, 221 kDa Homo sapiens neanderthalensis protein
|
| Buffer: |
10 mM HEPES, 100 mM NaCl, 1 mM DTT, 1 mM 5-aminoimidazole-4-carboxamide ribonucleotide (AICAR), 1mM Fumarate, pH: 7.5
|
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Jul 24
|
Molecular comparison of Neanderthal and Modern Human adenylosuccinate lyase.
Sci Rep 8(1):18008 (2018)
Van Laer B, Kapp U, Soler-Lopez M, Moczulska K, Pääbo S, Leonard G, Mueller-Dieckmann C
|
| RgGuinier |
3.6 |
nm |
| VolumePorod |
252 |
nm3 |
|
|
|
|
|
|
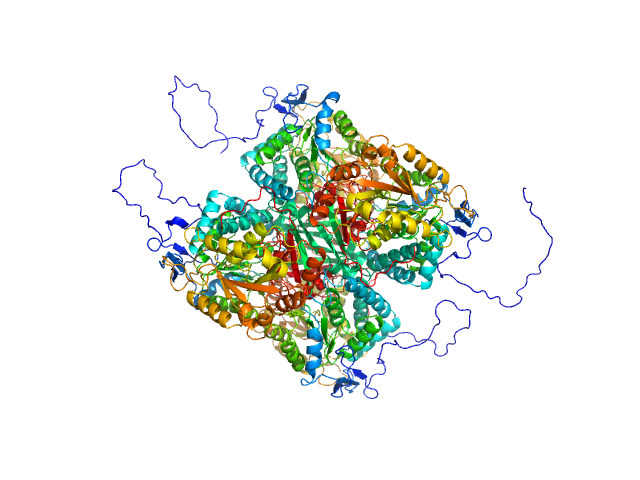
|
| Sample: |
Aldehyde dehydrogenase 12 tetramer, 242 kDa Zea mays protein
|
| Buffer: |
50 mM Tris-HCl, 50 mM NaCl, 0.5 mM TCEP, and 5% (v/v) glycerol, pH: 7.8
|
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2016 Dec 6
|
Structural and Biochemical Characterization of Aldehyde Dehydrogenase 12, the Last Enzyme of Proline Catabolism in Plants.
J Mol Biol (2018)
Korasick DA, Končitíková R, Kopečná M, Hájková E, Vigouroux A, Moréra S, Becker DF, Šebela M, Tanner JJ, Kopečný D
|
| RgGuinier |
4.1 |
nm |
| Dmax |
14.4 |
nm |
| VolumePorod |
351 |
nm3 |
|
|
|
|
|
|
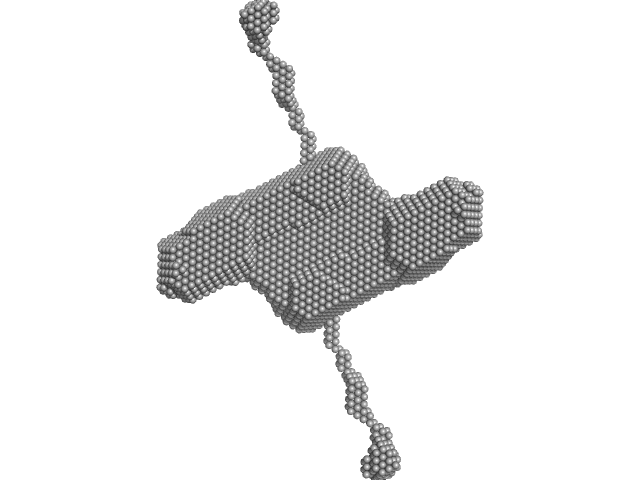
|
| Sample: |
Lipoprotein lipase dimer, 101 kDa Homo sapiens protein
Glycosylphosphatidylinositol-anchored high density lipoprotein-binding protein 1 dimer, 29 kDa Homo sapiens protein
|
| Buffer: |
10 mM BisTris, 150 mM NaCl, 4 mM CaCl2, 10% glycerol, pH: 7.4
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Dec 2
|
Structure of the lipoprotein lipase-GPIHBP1 complex that mediates plasma triglyceride hydrolysis.
Proc Natl Acad Sci U S A (2018)
Birrane G, Beigneux AP, Dwyer B, Strack-Logue B, Kristensen KK, Francone OL, Fong LG, Mertens HDT, Pan CQ, Ploug M, Young SG, Meiyappan M
|
| RgGuinier |
5.5 |
nm |
| Dmax |
19.0 |
nm |
| VolumePorod |
319 |
nm3 |
|
|
|
|
|
|
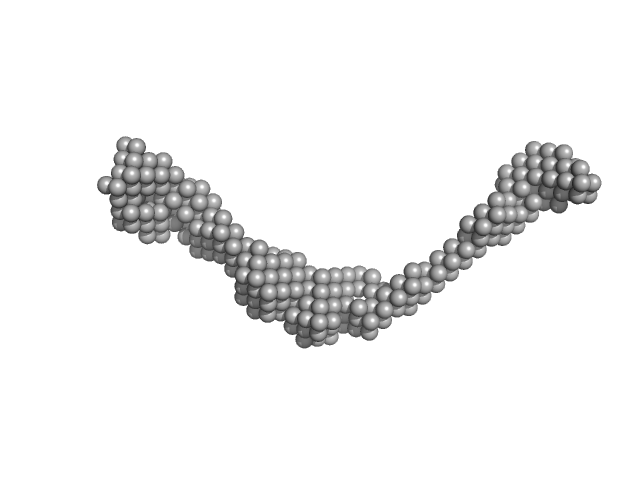
|
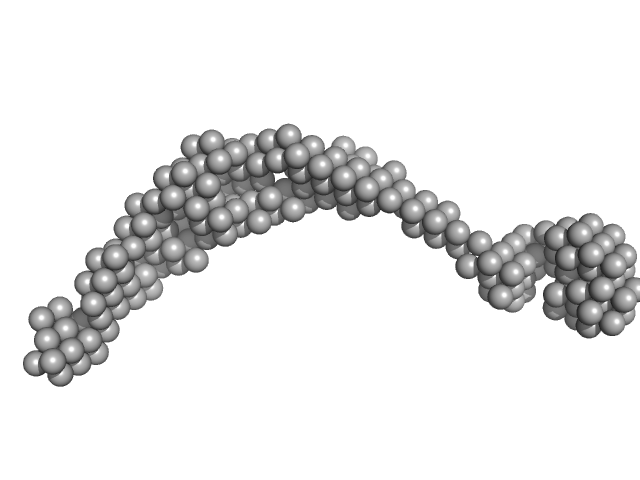
| Sample: |
Bromodomain-containing protein 4 monomer, 47 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 2% glycerol, 0.5 mM TCEP, pH: 7.5
|
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Jan 13
|
Interactome Rewiring Following Pharmacological Targeting of BET Bromodomains.
Mol Cell (2018)
Lambert JP, Picaud S, Fujisawa T, Hou H, Savitsky P, Uusküla-Reimand L, Gupta GD, Abdouni H, Lin ZY, Tucholska M, Knight JDR, Gonzalez-Badillo B, St-Denis N, Newman JA, Stucki M, Pelletier L, Bandeira...
|
| RgGuinier |
6.3 |
nm |
| Dmax |
22.0 |
nm |
| VolumePorod |
251 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Bromodomain-containing protein 3 monomer, 44 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 2% glycerol, 0.5 mM TCEP, pH: 7.5
|
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Jan 13
|
Interactome Rewiring Following Pharmacological Targeting of BET Bromodomains.
Mol Cell (2018)
Lambert JP, Picaud S, Fujisawa T, Hou H, Savitsky P, Uusküla-Reimand L, Gupta GD, Abdouni H, Lin ZY, Tucholska M, Knight JDR, Gonzalez-Badillo B, St-Denis N, Newman JA, Stucki M, Pelletier L, Bandeira...
|
| RgGuinier |
6.2 |
nm |
| Dmax |
21.5 |
nm |
| VolumePorod |
210 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Bromodomain-containing protein 2 monomer, 43 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 2% glycerol, 0.5 mM TCEP, pH: 7.5
|
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Jan 13
|
Interactome Rewiring Following Pharmacological Targeting of BET Bromodomains.
Mol Cell (2018)
Lambert JP, Picaud S, Fujisawa T, Hou H, Savitsky P, Uusküla-Reimand L, Gupta GD, Abdouni H, Lin ZY, Tucholska M, Knight JDR, Gonzalez-Badillo B, St-Denis N, Newman JA, Stucki M, Pelletier L, Bandeira...
|
| RgGuinier |
5.7 |
nm |
| Dmax |
21.0 |
nm |
| VolumePorod |
220 |
nm3 |
|
|
|
|
|
|

|
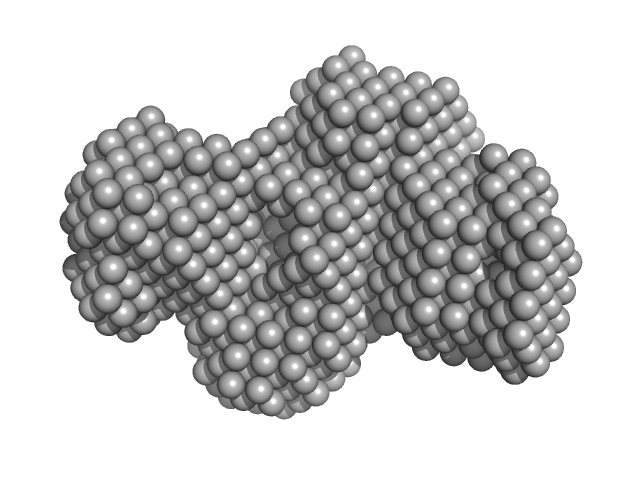
| Sample: |
Bromodomain testis-specific protein monomer, 43 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 2% glycerol, 0.5 mM TCEP, pH: 7.5
|
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Jan 13
|
Interactome Rewiring Following Pharmacological Targeting of BET Bromodomains.
Mol Cell (2018)
Lambert JP, Picaud S, Fujisawa T, Hou H, Savitsky P, Uusküla-Reimand L, Gupta GD, Abdouni H, Lin ZY, Tucholska M, Knight JDR, Gonzalez-Badillo B, St-Denis N, Newman JA, Stucki M, Pelletier L, Bandeira...
|
| RgGuinier |
5.1 |
nm |
| Dmax |
20.0 |
nm |
| VolumePorod |
200 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Gamma-crystallin S dimer, 42 kDa Homo sapiens protein
|
| Buffer: |
20 mM sodium phosphate, pH: 7
|
| Experiment: |
SAXS
data collected at Bruker Nanostar II, Australian Nuclear Science and Technology Organisation/Australian Centre for Neutron Scattering on 2018 Feb 23
|
The structure and stability of the disulfide-linked γS-crystallin dimer provide insight into oxidation products associated with lens cataract formation.
J Mol Biol (2018)
Thorn DC, Grosas AB, Mabbitt PD, Ray NJ, Jackson CJ, Carver JA
|
| RgGuinier |
2.4 |
nm |
| Dmax |
7.5 |
nm |
| VolumePorod |
45 |
nm3 |
|
|