|
|
|
|
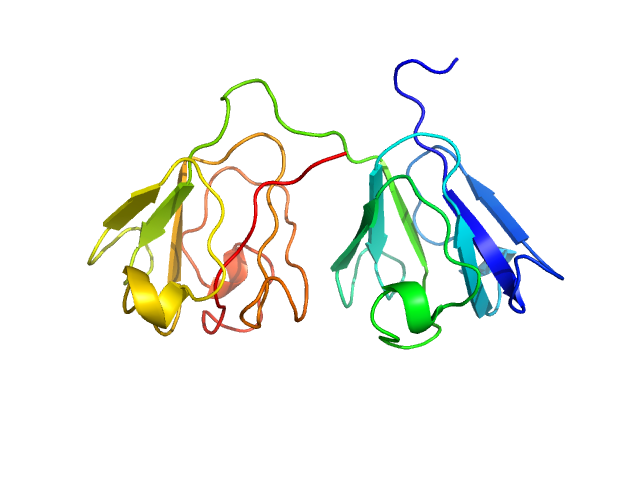
|
| Sample: |
Gamma-crystallin S monomer, 21 kDa Homo sapiens protein
|
| Buffer: |
20 mM sodium phosphate, pH: 7
|
| Experiment: |
SAXS
data collected at Bruker Nanostar II, Australian Nuclear Science and Technology Organisation/Australian Centre for Neutron Scattering on 2018 Feb 23
|
The structure and stability of the disulfide-linked γS-crystallin dimer provide insight into oxidation products associated with lens cataract formation.
J Mol Biol (2018)
Thorn DC, Grosas AB, Mabbitt PD, Ray NJ, Jackson CJ, Carver JA
|
| RgGuinier |
1.8 |
nm |
| Dmax |
5.9 |
nm |
| VolumePorod |
27 |
nm3 |
|
|
|
|
|
|
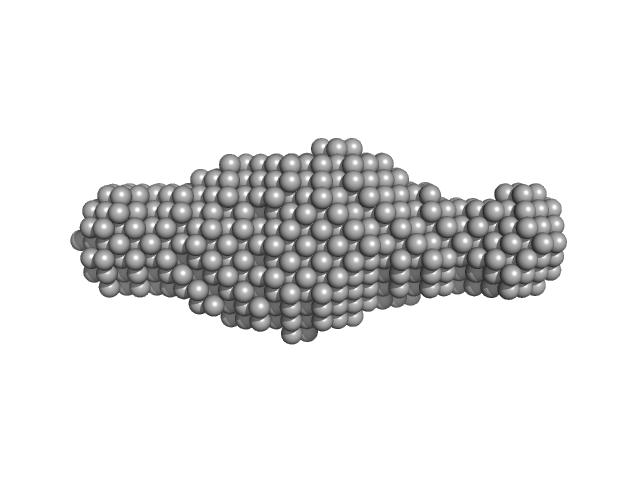
|
| Sample: |
Ribosome assembly protein 1 monomer, 124 kDa Saccharomyces cerevisiae protein
Ribosome maturation protein SDO1 monomer, 28 kDa Saccharomyces cerevisiae protein
|
| Buffer: |
50 mM Tris pH 8.0, 10% glycerol, 300 mM NaCl, 5 mM MgCl2., pH:
|
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Sep 21
|
Interaction of the GTPase Elongation Factor Like-1 with the Shwachman-Diamond Syndrome Protein and Its Missense Mutations.
Int J Mol Sci 19(12) (2018)
Gijsbers A, Montagut DC, Méndez-Godoy A, Altamura D, Saviano M, Siliqi D, Sánchez-Puig N
|
| RgGuinier |
5.0 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
333 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Ribosome assembly protein 1 monomer, 124 kDa Saccharomyces cerevisiae protein
|
| Buffer: |
50 mM Tris pH 8.0, 10% glycerol, 300 mM NaCl, 5 mM MgCl2., pH:
|
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Sep 21
|
Interaction of the GTPase Elongation Factor Like-1 with the Shwachman-Diamond Syndrome Protein and Its Missense Mutations.
Int J Mol Sci 19(12) (2018)
Gijsbers A, Montagut DC, Méndez-Godoy A, Altamura D, Saviano M, Siliqi D, Sánchez-Puig N
|
| RgGuinier |
4.7 |
nm |
| Dmax |
15.8 |
nm |
| VolumePorod |
258 |
nm3 |
|
|
|
|
|
|
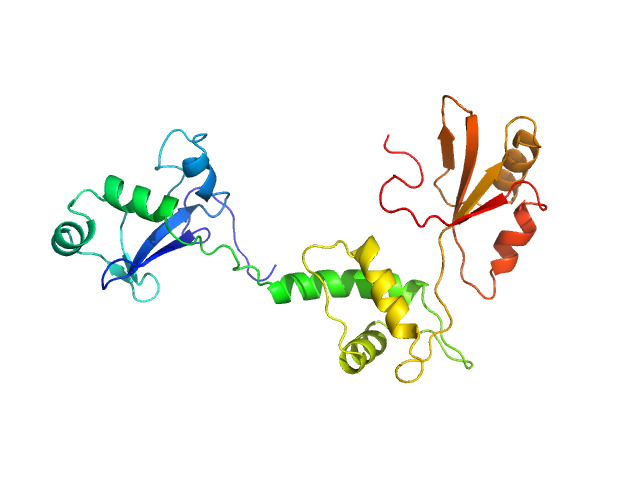
|
| Sample: |
Ribosome maturation protein SDO1 monomer, 28 kDa Saccharomyces cerevisiae protein
|
| Buffer: |
50 mM Tris pH 8.0, 10% glycerol, 300 mM NaCl, 5 mM MgCl2., pH:
|
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Sep 21
|
Interaction of the GTPase Elongation Factor Like-1 with the Shwachman-Diamond Syndrome Protein and Its Missense Mutations.
Int J Mol Sci 19(12) (2018)
Gijsbers A, Montagut DC, Méndez-Godoy A, Altamura D, Saviano M, Siliqi D, Sánchez-Puig N
|
| RgGuinier |
2.7 |
nm |
| Dmax |
8.5 |
nm |
|
|
|
|
|
|

|
| Sample: |
Ribosome maturation protein SDO1 monomer, 17 kDa Saccharomyces cerevisiae protein
|
| Buffer: |
50 mM Tris pH 8.0, 10% glycerol, 300 mM NaCl, 5 mM MgCl2., pH:
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2015 Aug 14
|
Interaction of the GTPase Elongation Factor Like-1 with the Shwachman-Diamond Syndrome Protein and Its Missense Mutations.
Int J Mol Sci 19(12) (2018)
Gijsbers A, Montagut DC, Méndez-Godoy A, Altamura D, Saviano M, Siliqi D, Sánchez-Puig N
|
| RgGuinier |
2.1 |
nm |
| Dmax |
6.2 |
nm |
| VolumePorod |
25 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Proliferating cell nuclear antigen trimer, 117 kDa Saccharomyces cerevisiae protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 5% glycerol, 1 mM TCEP, pH: 7.5
|
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2018 Aug 6
|
Conformational Flexibility of Ubiquitin-Modified and SUMO-Modified PCNA Shown by Full-Ensemble Hybrid Methods.
J Mol Biol 430(24):5294-5303 (2018)
Powers KT, Lavering ED, Washington MT
|
| RgGuinier |
4.1 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
238 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Proliferating cell nuclear antigen trimer, 125 kDa Saccharomyces cerevisiae protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 5% glycerol, 1 mM TCEP, pH: 7.5
|
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2018 Aug 6
|
Conformational Flexibility of Ubiquitin-Modified and SUMO-Modified PCNA Shown by Full-Ensemble Hybrid Methods.
J Mol Biol 430(24):5294-5303 (2018)
Powers KT, Lavering ED, Washington MT
|
| RgGuinier |
4.7 |
nm |
| Dmax |
16.9 |
nm |
| VolumePorod |
310 |
nm3 |
|
|
|
|
|
|
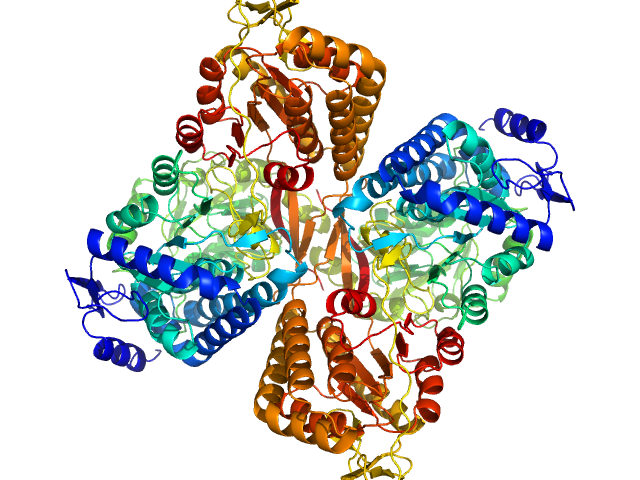
|
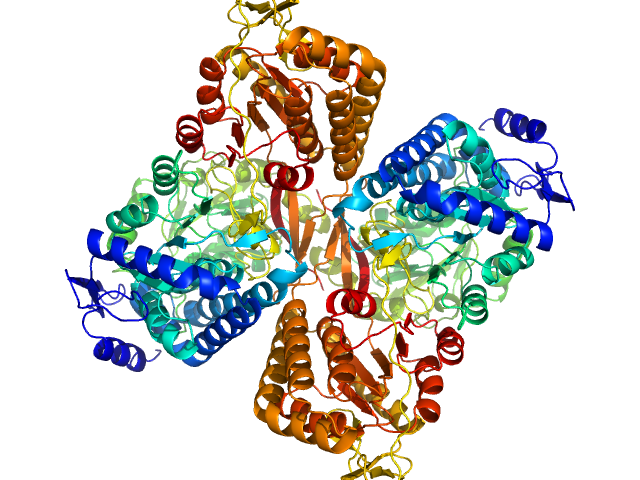
| Sample: |
Aldehyde dehydrogenase 16 from Loktanella sp. dimer, 161 kDa Loktanella sp. 3ANDIMAR09 protein
|
| Buffer: |
20 mM Tris-HCl, 100 mM NaCl, 2.0% glycerol, 0.5 mM Tris(3-hydroxypropyl)phosphine, pH: 8
|
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2017 Dec 13
|
Crystal Structure of Aldehyde Dehydrogenase 16 Reveals Trans-Hierarchical Structural Similarity and a New Dimer.
J Mol Biol (2018)
Liu LK, Tanner JJ
|
| RgGuinier |
3.6 |
nm |
| Dmax |
10.9 |
nm |
| VolumePorod |
202 |
nm3 |
|
|
|
|
|
|
|
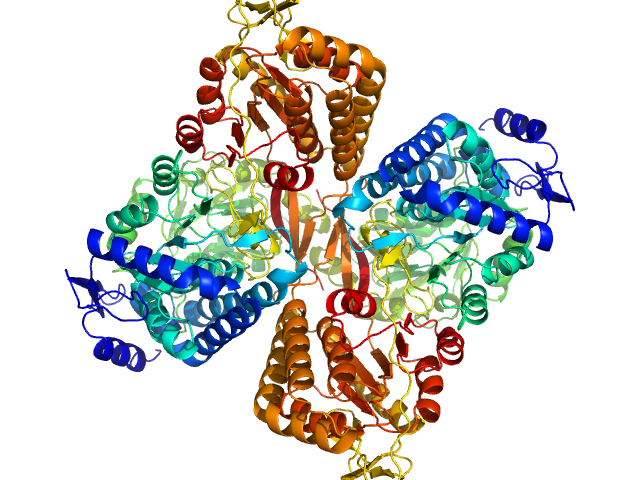
| Sample: |
Aldehyde dehydrogenase 16 from Loktanella sp. dimer, 161 kDa Loktanella sp. 3ANDIMAR09 protein
|
| Buffer: |
20 mM Tris-HCl, 100 mM NaCl, 2.0% glycerol, 0.5 mM Tris(3-hydroxypropyl)phosphine, pH: 8
|
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2017 Dec 13
|
Crystal Structure of Aldehyde Dehydrogenase 16 Reveals Trans-Hierarchical Structural Similarity and a New Dimer.
J Mol Biol (2018)
Liu LK, Tanner JJ
|
| RgGuinier |
3.6 |
nm |
| Dmax |
11.2 |
nm |
| VolumePorod |
204 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Aldehyde dehydrogenase 16 from Loktanella sp. dimer, 161 kDa Loktanella sp. 3ANDIMAR09 protein
|
| Buffer: |
20 mM Tris-HCl, 100 mM NaCl, 2.0% glycerol, 0.5 mM Tris(3-hydroxypropyl)phosphine, pH: 8
|
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2017 Dec 13
|
Crystal Structure of Aldehyde Dehydrogenase 16 Reveals Trans-Hierarchical Structural Similarity and a New Dimer.
J Mol Biol (2018)
Liu LK, Tanner JJ
|
| RgGuinier |
3.5 |
nm |
| Dmax |
10.6 |
nm |
| VolumePorod |
207 |
nm3 |
|
|