|
|
|
|
|
| Sample: |
Probable esterase KAI2 - H246Y (KARRIKIN INSENSITIVE 2 - H246Y) monomer, 30 kDa Arabidopsis thaliana protein
|
| Buffer: |
20 mM HEPES, 150 mM imidazole, 150 mM NaCl, 10% (v/v) glycerol, pH: 7.5
|
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2022 Mar 29
|
SAXS studies of KAI2 catalytic mutants
Sabrina Davies
|
| RgGuinier |
3.0 |
nm |
| Dmax |
16.1 |
nm |
| VolumePorod |
70 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Probable esterase KAI2 - H246Y (KARRIKIN INSENSITIVE 2 - H246Y) monomer, 30 kDa Arabidopsis thaliana protein
|
| Buffer: |
20 mM HEPES, 150 mM imidazole, 150 mM NaCl, 10% (v/v) glycerol, pH: 7.5
|
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2022 Mar 29
|
SAXS studies of KAI2 catalytic mutants
Sabrina Davies
|
| RgGuinier |
2.9 |
nm |
| Dmax |
13.2 |
nm |
| VolumePorod |
68 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Probable esterase KAI2 - H246Y (KARRIKIN INSENSITIVE 2 - H246Y) monomer, 30 kDa Arabidopsis thaliana protein
|
| Buffer: |
20 mM HEPES, 150 mM imidazole, 150 mM NaCl, 10% (v/v) glycerol, pH: 7.5
|
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2022 Mar 29
|
SAXS studies of KAI2 catalytic mutants
Sabrina Davies
|
| RgGuinier |
5.6 |
nm |
| Dmax |
23.0 |
nm |
| VolumePorod |
494 |
nm3 |
|
|
|
|
|
|
|
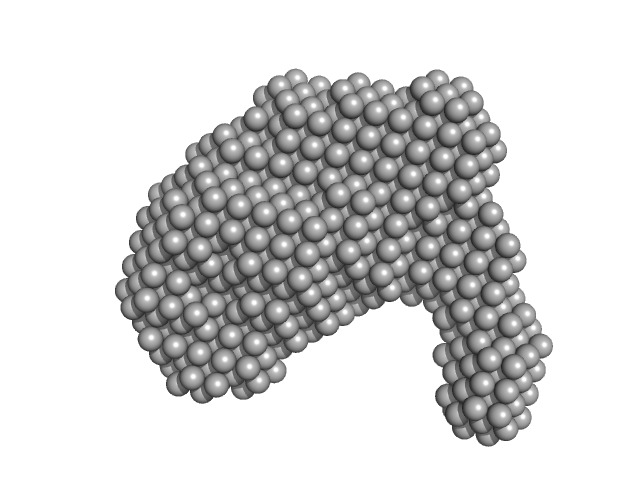
| Sample: |
Probable esterase KAI2 - H246Y (KARRIKIN INSENSITIVE 2 - H246Y) monomer, 30 kDa Arabidopsis thaliana protein
|
| Buffer: |
20 mM HEPES, 150 mM imidazole, 150 mM NaCl, 10% (v/v) glycerol, pH: 7.5
|
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2022 Mar 29
|
SAXS studies of KAI2 catalytic mutants
Sabrina Davies
|
| RgGuinier |
5.1 |
nm |
| Dmax |
19.3 |
nm |
| VolumePorod |
403 |
nm3 |
|
|
|
|
|
|
|
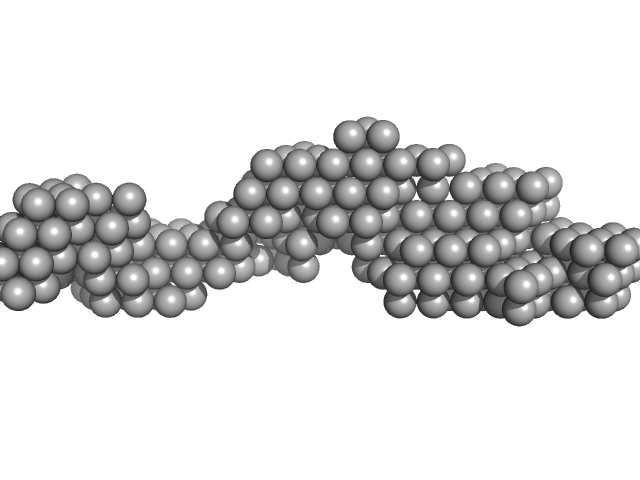
| Sample: |
ESX-5 secretion-associated protein EspG5 monomer, 32 kDa Mycobacterium tuberculosis (strain … protein
ESX-5 secretion system protein EccA5 monomer, 31 kDa Mycobacterium tuberculosis (strain … protein
|
| Buffer: |
50 mM HEPES, 200 mM NaCl, pH: 8
|
| Experiment: |
SAXS
data collected at Anton Paar SAXSpace, CSIR-Central Drug Research Institute on 2025 Jan 28
|
Small angle x-ray scattering envelope of EccA5 N-terminal domain and EspG5 of Type 7 secretion pathway of Mycobacterium tuberculosis
Rajlakshmi K
|
| RgGuinier |
3.5 |
nm |
| Dmax |
9.1 |
nm |
| VolumePorod |
83 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
BC120 RNA monomer monomer, 39 kDa RNA
|
| Buffer: |
phosphate buffered saline, pH: 7.4
|
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2023 Sep 22
|
Alu RNA pseudoknot alterations influence SRP9/SRP14 association.
RNA (2025)
Gussakovsky D, Brown MJF, Pereira HS, Meier M, Padilla-Meier GP, Black NA, Booy EP, Stetefeld J, Patel TR, McKenna SA
|
| RgGuinier |
4.0 |
nm |
| Dmax |
14.3 |
nm |
| VolumePorod |
57 |
nm3 |
|
|
|
|
|
|

|
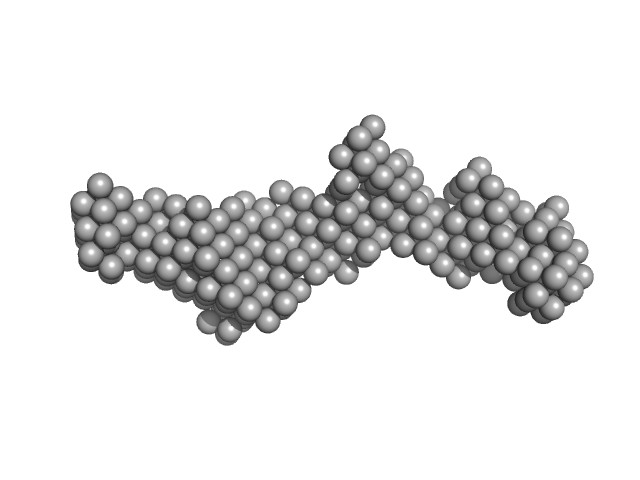
| Sample: |
EB120 RNA monomer monomer, 39 kDa RNA
|
| Buffer: |
phosphate buffered saline, pH: 7.4
|
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2023 Sep 22
|
Alu RNA pseudoknot alterations influence SRP9/SRP14 association.
RNA (2025)
Gussakovsky D, Brown MJF, Pereira HS, Meier M, Padilla-Meier GP, Black NA, Booy EP, Stetefeld J, Patel TR, McKenna SA
|
| RgGuinier |
4.2 |
nm |
| Dmax |
13.6 |
nm |
| VolumePorod |
79 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
BC120 G25C RNA monomer, 39 kDa Homo sapiens RNA
|
| Buffer: |
phosphate buffered saline, pH: 7.4
|
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2023 Sep 22
|
Alu RNA pseudoknot alterations influence SRP9/SRP14 association.
RNA (2025)
Gussakovsky D, Brown MJF, Pereira HS, Meier M, Padilla-Meier GP, Black NA, Booy EP, Stetefeld J, Patel TR, McKenna SA
|
| RgGuinier |
4.1 |
nm |
| Dmax |
13.8 |
nm |
| VolumePorod |
63 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Aromatic-L-amino-acid decarboxylase (L353P) dimer, 107 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, pH: 7.4
|
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2021 Jul 7
|
The CRISPR-Cas9 knockout DDC SH-SY5Y in vitro model for AADC deficiency provides insight into the pathogenicity of R347Q and L353P variants: a cross-sectional structural and functional analysis.
FEBS J (2025)
Carmona-Carmona CA, Bisello G, Franchini R, Lunardi G, Galavotti R, Perduca M, Ribeiro RP, Belviso BD, Giorgetti A, Caliandro R, Lievens PM, Bertoldi M
|
| RgGuinier |
3.1 |
nm |
| Dmax |
8.8 |
nm |
| VolumePorod |
152 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Aromatic-L-amino-acid decarboxylase (R347Q) dimer, 108 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, 100 µM pyridoxal 5'-phosphate, pH: 7.4
|
| Experiment: |
SAXS
data collected at BM29, ESRF on 2021 Jul 7
|
The CRISPR-Cas9 knockout DDC SH-SY5Y in vitro model for AADC deficiency provides insight into the pathogenicity of R347Q and L353P variants: a cross-sectional structural and functional analysis.
FEBS J (2025)
Carmona-Carmona CA, Bisello G, Franchini R, Lunardi G, Galavotti R, Perduca M, Ribeiro RP, Belviso BD, Giorgetti A, Caliandro R, Lievens PM, Bertoldi M
|
| RgGuinier |
3.6 |
nm |
| Dmax |
14.2 |
nm |
| VolumePorod |
220 |
nm3 |
|
|