|
|
|
|
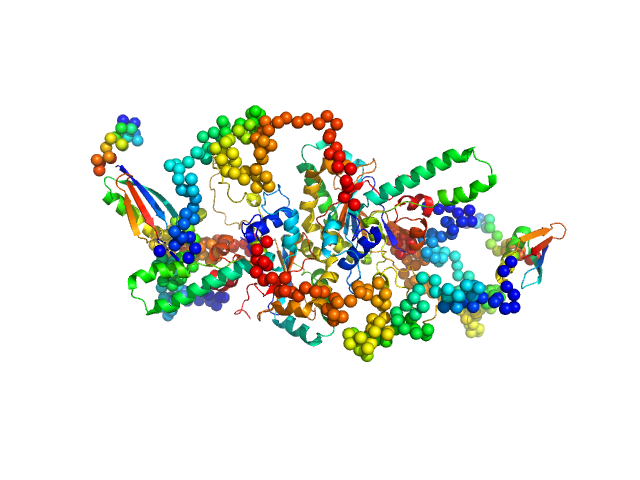
|
| Sample: |
Uncharacterized protein dimer, 122 kDa Ustilago maydis (strain … protein
|
| Buffer: |
20mM HEPES, 20 mM KCl, 200 mM NaCl, pH: 7.5
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2023 Oct 25
|
Specific phosphoinositide interaction of Jps1 is a key feature during unconventional secretion in Ustilago maydis
Journal of Biological Chemistry :110215 (2025)
Dali S, Schultz M, Köster M, Kamel M, Busch M, Steinchen W, Hänsch S, Papadopoulos A, Reiners J, Smits S, Kedrov A, Altegoer F, Schipper K
|
| RgGuinier |
4.0 |
nm |
| Dmax |
14.7 |
nm |
| VolumePorod |
254 |
nm3 |
|
|
|
|
|
|
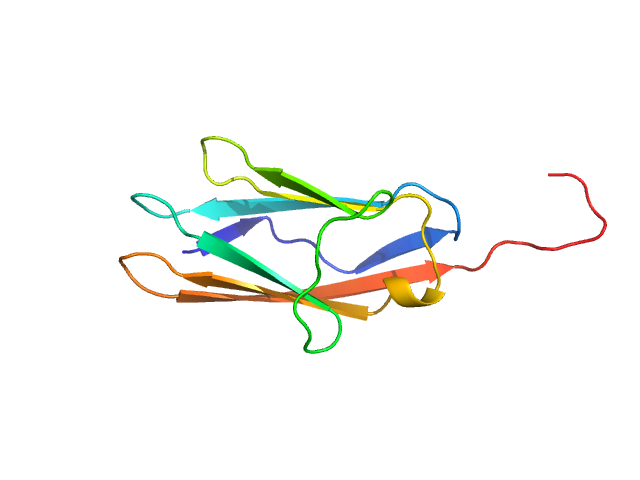
|
| Sample: |
Palladin monomer, 12 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES pH 7.4, 1 mM DTT, 100 mM NaCl, pH:
|
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2023 Aug 29
|
Integrated structural model of the palladin-actin complex using XL-MS, docking, NMR, and SAXS.
Protein Sci 34(5):e70122 (2025)
Sargent R, Liu DH, Yadav R, Glennenmeier D, Bradford C, Urbina N, Beck MR
|
| RgGuinier |
1.7 |
nm |
| Dmax |
6.8 |
nm |
| VolumePorod |
18 |
nm3 |
|
|
|
|
|
|
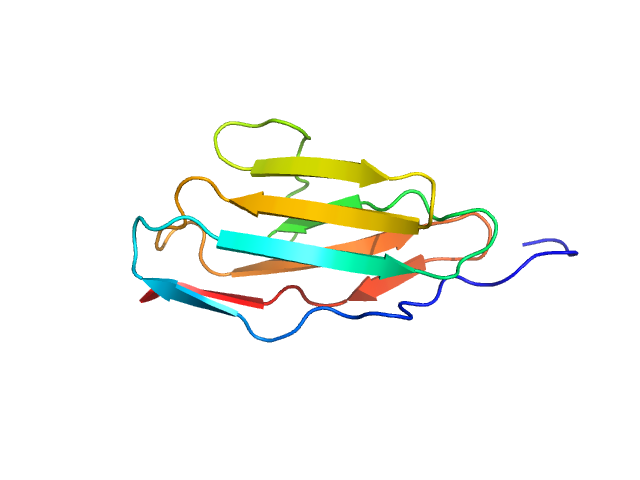
|
| Sample: |
Palladin monomer, 12 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES pH 7.4, 1 mM DTT, 100 mM NaCl, pH:
|
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2023 Aug 29
|
Integrated structural model of the palladin-actin complex using XL-MS, docking, NMR, and SAXS.
Protein Sci 34(5):e70122 (2025)
Sargent R, Liu DH, Yadav R, Glennenmeier D, Bradford C, Urbina N, Beck MR
|
| RgGuinier |
1.6 |
nm |
| Dmax |
6.7 |
nm |
| VolumePorod |
18 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Palladin monomer, 27 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES pH 7.4, 1 mM DTT, 100 mM NaCl, pH:
|
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2023 Aug 29
|
Integrated structural model of the palladin-actin complex using XL-MS, docking, NMR, and SAXS.
Protein Sci 34(5):e70122 (2025)
Sargent R, Liu DH, Yadav R, Glennenmeier D, Bradford C, Urbina N, Beck MR
|
| RgGuinier |
2.8 |
nm |
| Dmax |
12.3 |
nm |
| VolumePorod |
29 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Cadherin EGF LAG seven-pass G-type receptor 1 monomer, 245 kDa Mus musculus protein
|
| Buffer: |
10 mM Tris, 150 mM NaCl, pH: 8.5
|
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Dec 8
|
Structural basis for regulation of CELSR1 by a compact module in its extracellular region.
Nat Commun 16(1):3972 (2025)
Bandekar SJ, Garbett K, Kordon SP, Dintzner EE, Li J, Shearer T, Sando RC, Araç D
|
| RgGuinier |
6.0 |
nm |
| Dmax |
22.5 |
nm |
| VolumePorod |
473 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Cadherin EGF LAG seven-pass G-type receptor 1 dimer, 491 kDa Mus musculus protein
|
| Buffer: |
10 mM Tris, 150 mM NaCl, 1 mM CaCl2, pH: 8.5
|
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Dec 8
|
Structural basis for regulation of CELSR1 by a compact module in its extracellular region.
Nat Commun 16(1):3972 (2025)
Bandekar SJ, Garbett K, Kordon SP, Dintzner EE, Li J, Shearer T, Sando RC, Araç D
|
| RgGuinier |
16.2 |
nm |
| Dmax |
67.5 |
nm |
| VolumePorod |
722 |
nm3 |
|
|
|
|
|
|
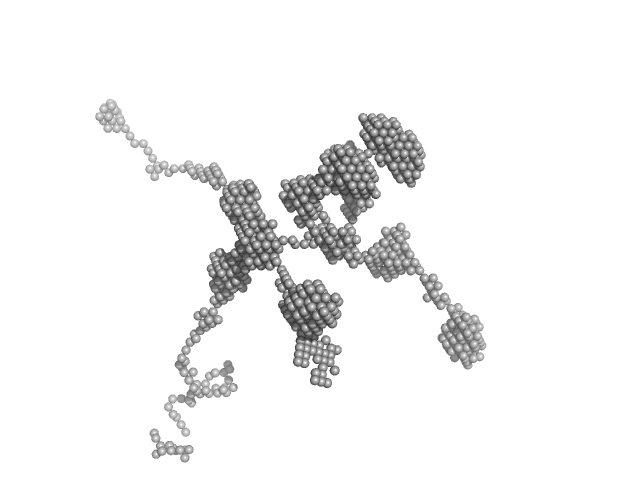
|
| Sample: |
self-amplifying RNA monomer, 3030 kDa RNA
|
| Buffer: |
MBG buffer: 5% w/v D-Glucose, 10mM MES (2-(N-morpholino)ethanesulfonic acid) in double distillated sterile/RNAse free water, pH: 6.1
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Nov 10
|
Compact polyethylenimine-complexed mRNA molecules as quintessential vaccines
Martin Schroer
|
| RgGuinier |
90.5 |
nm |
| Dmax |
200.0 |
nm |
|
|
|
|
|
|

|
| Sample: |
self-amplifying RNA monomer, 3030 kDa RNA
|
| Buffer: |
MBG buffer: 5% w/v D-Glucose, 10mM MES (2-(N-morpholino)ethanesulfonic acid) in double distillated sterile/RNAse free water, pH: 6.1
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Mar 10
|
Compact polyethylenimine-complexed mRNA molecules as quintessential vaccines
Martin Schroer
|
| RgGuinier |
30.7 |
nm |
| Dmax |
107.4 |
nm |
| VolumePorod |
112000 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
self-amplifying RNA monomer, 3030 kDa RNA
linear polyethylenimine monomer, 25 kDa none (polymer)
|
| Buffer: |
MBG buffer: 5% w/v D-Glucose, 10mM MES (2-(N-morpholino)ethanesulfonic acid) in double distillated sterile/RNAse free water, pH: 6.1
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Nov 10
|
Compact polyethylenimine-complexed mRNA molecules as quintessential vaccines
Martin Schroer
|
| RgGuinier |
12.0 |
nm |
| Dmax |
40.0 |
nm |
| VolumePorod |
9100 |
nm3 |
|
|
|
|
|
|
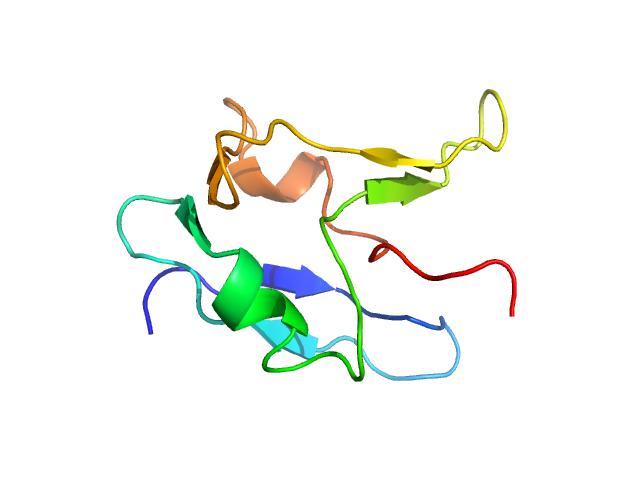
|
| Sample: |
TAR DNA-binding protein 43 monomer, 9 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris-HCl, 150 mM NaCl, 1 mM DTT, 3% glycerol, pH: 8.5
|
| Experiment: |
SAXS
data collected at BL38B1, SPring-8 on 2024 May 29
|
Semi-synthesis of TDP-43 reveals the effects of phosphorylation in N-terminal domain on self-association
Communications Chemistry 8(1) (2025)
Sasaki D, Tenda M, Sohma Y
|
| RgGuinier |
1.4 |
nm |
| Dmax |
4.3 |
nm |
| VolumePorod |
15 |
nm3 |
|
|