Structure of the PCBP2/stem-loop IV complex underlying translation initiation mediated by the poliovirus type I IRES.
Beckham SA,
Matak MY,
Belousoff MJ,
Venugopal H,
Shah N,
Vankadari N,
Elmlund H,
Nguyen JHC,
Semler BL,
Wilce MCJ,
Wilce JA
Nucleic Acids Res
(2020 Jun 18)
|
|
|
|
|
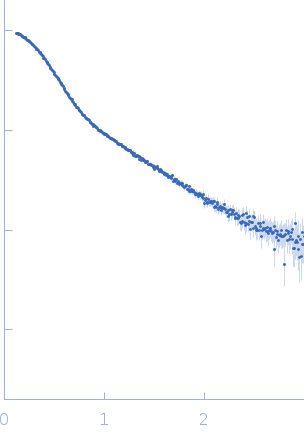
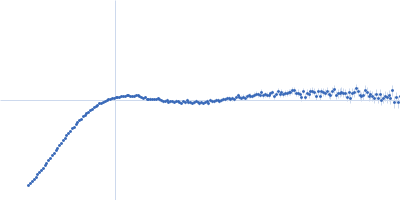
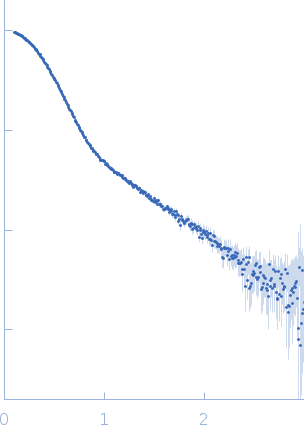
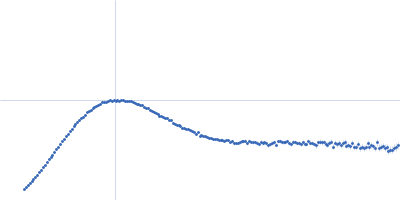
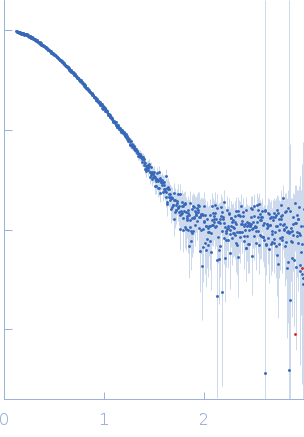
| Sample: |
Modified stem loop IV poliovirus IRES, nucleotides 278-398 monomer, 41 kDa Human poliovirus 1 … RNA
|
| Buffer: |
5 mM HEPES-KOH, 25 mM KCl, 2 mM MgCl2, 2 mM DTT, 4 % glycerol, 0.1 mM EDTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2017 Dec 2
|
|
| RgGuinier |
3.5 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
94 |
nm3 |
|
|
|
|
|
|
|
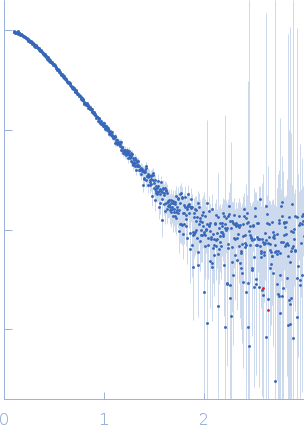
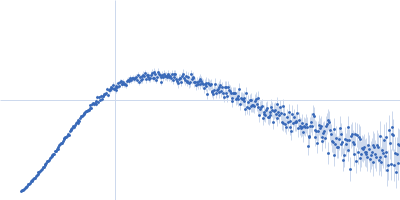
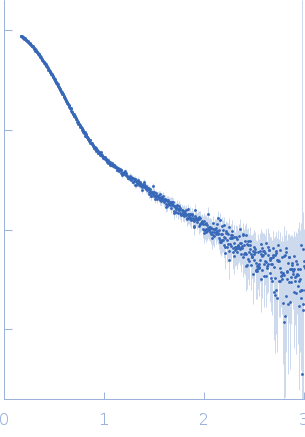
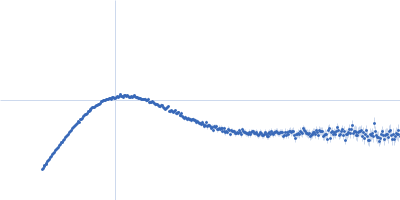
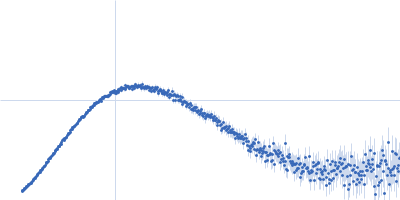
| Sample: |
Poly(rC)-binding protein 2 monomer, 40 kDa Homo sapiens protein
|
| Buffer: |
5 mM HEPES-KOH, 25 mM KCl, 2 mM MgCl2, 2 mM DTT, 4 % glycerol, 0.1 mM EDTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2017 Jul 16
|
|
| RgGuinier |
3.3 |
nm |
| Dmax |
13.2 |
nm |
| VolumePorod |
76 |
nm3 |
|
|
|
|
|
|
|
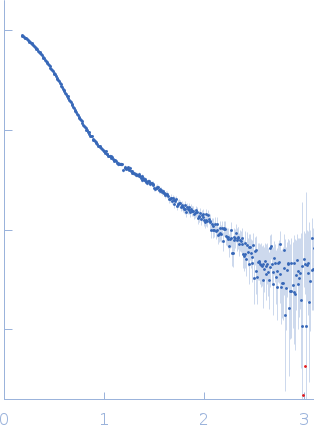
| Sample: |
Poly(rC)-binding protein 2 monomer, 40 kDa Homo sapiens protein
Modified stem loop IV poliovirus IRES, nucleotides 278-398 monomer, 41 kDa Human poliovirus 1 … RNA
|
| Buffer: |
5 mM HEPES-KOH, 25 mM KCl, 2 mM MgCl2, 2 mM DTT, 4 % glycerol, 0.1 mM EDTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2017 Dec 16
|
|
| RgGuinier |
3.7 |
nm |
| Dmax |
11.5 |
nm |
| VolumePorod |
162 |
nm3 |
|
|
|
|
|
|
|
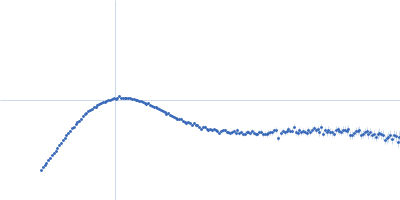
| Sample: |
Modified stem loop IV poliovirus IRES, nucleotides 278-398 monomer, 41 kDa Human poliovirus 1 … RNA
Truncated poly(rC)-binding protein 2 (ΔKH3) monomer, 28 kDa Homo sapiens protein
Truncated poly(rC)-binding protein 2 (ΔKH3) monomer, 28 kDa Homo sapiens protein
|
| Buffer: |
5 mM HEPES-KOH, 25 mM KCl, 2 mM MgCl2, 2 mM DTT, 4 % glycerol, 0.1 mM EDTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2017 Jul 1
|
|
| RgGuinier |
3.8 |
nm |
| Dmax |
12.2 |
nm |
| VolumePorod |
165 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Truncated poly(rC)-binding protein 2 (ΔKH3) monomer, 28 kDa Homo sapiens protein
|
| Buffer: |
5 mM HEPES-KOH, 25 mM KCl, 2 mM MgCl2, 2 mM DTT, 4 % glycerol, 0.1 mM EDTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2017 Jan 1
|
|
| RgGuinier |
2.6 |
nm |
| Dmax |
9.2 |
nm |
| VolumePorod |
66 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Modified stem loop IV poliovirus IRES, nucleotides 278-398 monomer, 41 kDa Human poliovirus 1 … RNA
Truncated poly(rC)-binding protein 2 (ΔKH1-KH2) monomer, 18 kDa Homo sapiens protein
|
| Buffer: |
5 mM HEPES-KOH, 25 mM KCl, 2 mM MgCl2, 2 mM DTT, 4 % glycerol, 0.1 mM EDTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2017 Jul 1
|
|
| RgGuinier |
3.5 |
nm |
| Dmax |
12.2 |
nm |
| VolumePorod |
126 |
nm3 |
|
|