|
|
|
|
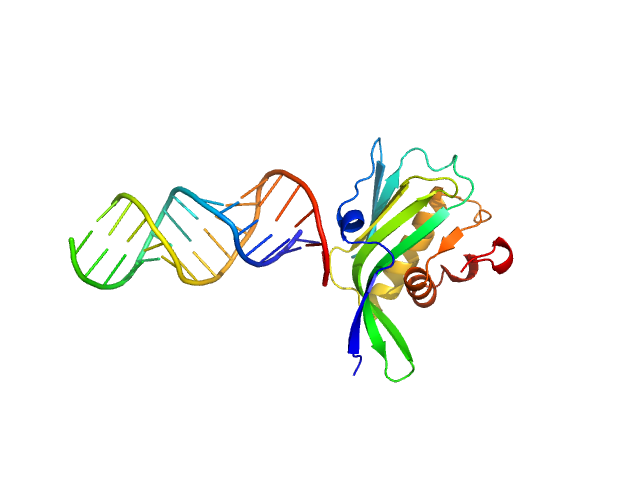
|
| Sample: |
Non-structural protein 1 monomer, 16 kDa Influenza B virus protein
RNA top strand monomer, 5 kDa RNA
RNA bottom strand monomer, 5 kDa RNA
|
| Buffer: |
40 mM ammonium acetate, pH 5.5, 450 mM NaCl, 5 mM CaCl2, 0.02% NaN3, 50 mM arginine, pH: 5.5
|
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2017 May 18
|
The NS1 protein of influenza B virus binds 5’-triphosphorylated dsRNA to suppress RIG-I activation and the host antiviral response
(2024)
...Tsutakawa S, Zhao C, Ma L, Shurina B, Hura G, John R, Vorobiev S, Swapna G, Solotchi M, Tainer J, Krug R, Patel S, Montelione G
|
| RgGuinier |
2.8 |
nm |
| Dmax |
10.3 |
nm |
| VolumePorod |
23 |
nm3 |
|