|
|
|
|
|
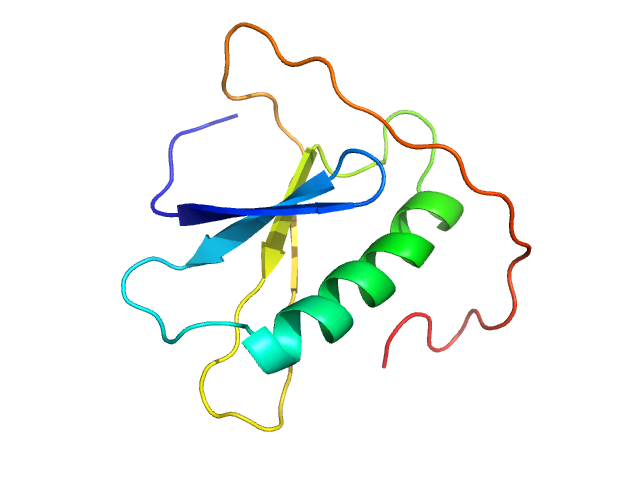
| Sample: |
Cholera toxin transcriptional activator monomer, 12 kDa Vibrio cholerae serotype … protein
|
| Buffer: |
50 mM Na2HPO4, 300 mM NaCl, 3% glycerol, pH: 8
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2022 Mar 6
|
Vibrio cholerae's ToxRS bile sensing system.
Elife 12 (2023)
...Gräwert MA, Rotzinger M, Berger TMI, Schäfer J, Usón I, Reidl J, Sánchez-Murcia PA, Zangger K, Pavkov-Keller T
|
| RgGuinier |
1.6 |
nm |
| Dmax |
6.0 |
nm |
| VolumePorod |
16 |
nm3 |
|