|
|
|
|
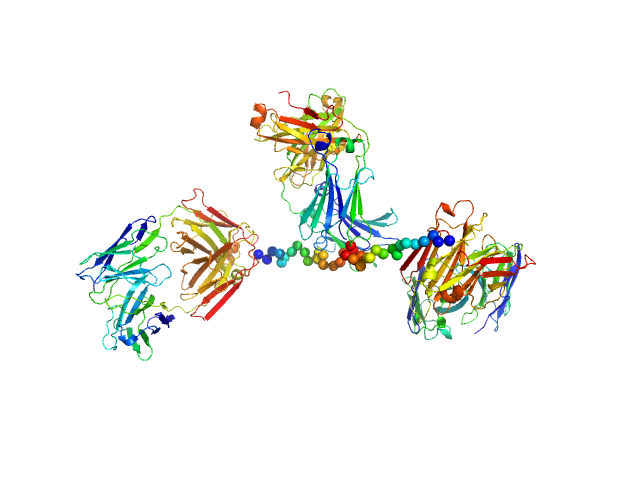
|
| Sample: |
Anti-prion protein monoclonal IgG2a 6D11 monomer, 145 kDa Mus musculus protein
|
| Buffer: |
phosphate buffered saline, pH: 7.4
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Dec 15
|
Ligands binding to the prion protein induce its proteolytic release with therapeutic potential in neurodegenerative proteinopathies.
Sci Adv 7(48):eabj1826 (2021)
...Damme M, Song F, Schwarz A, Da Vela S, Massignan T, Jung S, Correia A, Schmitz M, Puig B, Hornemann S, Zerr I, Tatzelt J, Biasini E, Saftig P, Schweizer M, Svergun D, Amin L, Mazzola F, Varani L, Thap...
|
| RgGuinier |
5.1 |
nm |
| Dmax |
17.4 |
nm |
| VolumePorod |
330 |
nm3 |
|