|
|
|
|
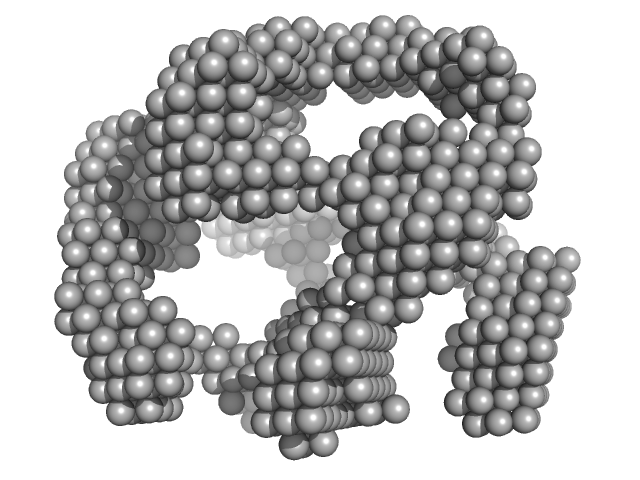
|
| Sample: |
lysozyme amyloid fibril , 1 kDa Gallus gallus protein
Fe3O4 nanoparticles; nominal diameter 20 nm (hydrodynamic diameter) monomer, 1 kDa
|
| Buffer: |
0.2 M glycine-HCl, 80 mM NaCl, pH: 2.2
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Sep 2
|
Dependence of the Nanoscale Composite Morphology of Fe3O4 Nanoparticle-Infused Lysozyme Amyloid Fibrils on Timing of Infusion: A Combined SAXS and AFM Study
Molecules 26(16):4864 (2021)
Schroer M, Hu P, Tomasovicova N, Batkova M, Zakutanska K, Wu P, Kopcansky P
|
| RgGuinier |
31.0 |
nm |
| Dmax |
75.0 |
nm |
|