|
Synchrotron SAXS
data from solutions of
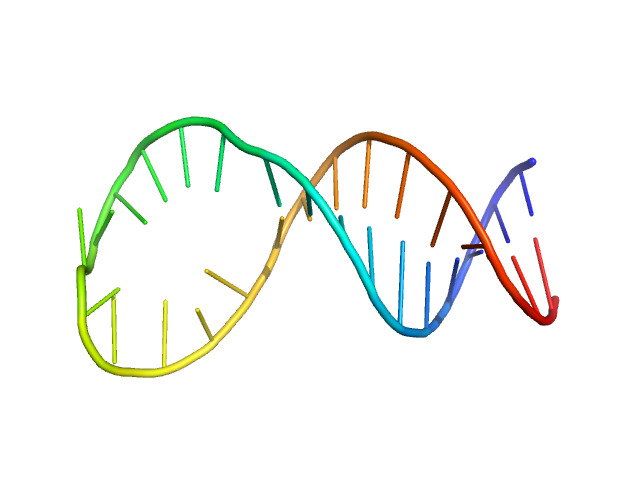
Apt31 - ssDNA aptamer specific to the receptor-binding domain of SARS-CoV-2
in
Tris-HCl, pH 7.4
were collected
on the
13A beam line
at the Taiwan Photon Source, NSRRC storage ring
(Hsinchu, Taiwan)
using a Eiger X 9M detector
at a sample-detector distance of 1.2 m and
at a wavelength of λ = 0.0827 nm
(I(s) vs s, where s = 4πsinθ/λ, and 2θ is the scattering angle).
In-line size-exclusion chromatography (SEC) SAS was employed. The SEC parameters were as follows: A sample
at 13.4 mg/ml was injected
onto a column
at 22°C.
136 successive
2 second frames were collected.
The data were normalized to the intensity of the transmitted beam and radially averaged; the scattering of the solvent-blank was subtracted.
To reconstruct the spatial structure of the Apt31 aptamer, the size-exclusion-chromatography (SEC) SAXS measurements were performed at the TPS 13A BioSAXS beamline at NSRRC, Taiwan. The sample was prepared in Tris buffer solution and exposed by X-rays of 15 keV at 22°C with an online HPLC system. The initial concentration was 13.4 mg/ml, HPLC column provided a high dilution of the sample during the measurements. The X-Ray beam size at the sample position was 320x260 μm². Sample to detector distance was 1210.06 mm. Total 128 data frames were recorded by the detector Eiger X 9M, each 2s exposure time per frame. All of the frames recorded with a constant Rg value over the HPLC (high-performance liquid chromatography) sample flow were radially averaged into the one-dimensional SAXS curve, two of them were merged into the resulted SAXS curve.
|
|
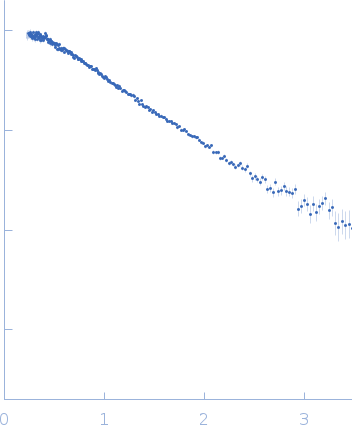 s, nm-1
s, nm-1