UniProt ID: Q9UHC6 (1-1261) Contactin-associated protein-like 2 extracellular domains (1-1261)
|
|
|

|
| Sample: |
Contactin-associated protein-like 2 extracellular domains (1-1261) monomer, 140 kDa Homo sapiens protein
|
| Buffer: |
10 mM HEPES 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSess, University of Utah on 2010 Oct 4
|
Structural Characterization of the Extracellular Domain of CASPR2 and Insights into Its Association with the Novel Ligand Contactin1.
J Biol Chem 291(11):5788-802 (2016)
Rubio-Marrero EN, Vincelli G, Jeffries CM, Shaikh TR, Pakos IS, Ranaivoson FM, von Daake S, Demeler B, De Jaco A, Perkins G, Ellisman MH, Trewhella J, Comoletti D
|
| RgGuinier |
4.4 |
nm |
| Dmax |
14.5 |
nm |
| VolumePorod |
282 |
nm3 |
|
|
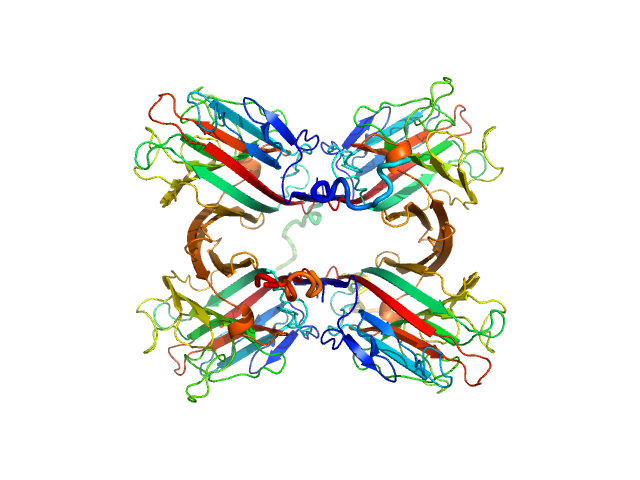
UniProt ID: P81371 (None-None) Recombinant Tn antigen-binding lectin
|
|
|
|
| Sample: |
Recombinant Tn antigen-binding lectin tetramer, 105 kDa Vatairea macrocarpa protein
|
| Buffer: |
100 mM sodium phosphate 150 mM NaCl 5% (v/v) glycerol, pH: 5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2014 Jan 11
|
Structural characterization of a Vatairea macrocarpa lectin in complex with a tumor-associated antigen: A new tool for cancer research.
Int J Biochem Cell Biol 72:27-39 (2016)
Sousa BL, Silva-Filho JC, Kumar P, Graewert MA, Pereira RI, Cunha RMS, Nascimento KS, Bezerra GA, Delatorre P, Djinovic-Carugo K, Nagano CS, Gruber K, Cavada BS
|
| RgGuinier |
3.2 |
nm |
| Dmax |
9.5 |
nm |
| VolumePorod |
168 |
nm3 |
|
|
UniProt ID: Q9H2K2 (489-649) Ankyrin repeat domains from Tankyrase 2
|
|
|
|
| Sample: |
Ankyrin repeat domains from Tankyrase 2 monomer, 18 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES 100mM NaCl 1mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2014 Dec 4
|
Hybrid Structural Analysis of the Arp2/3 Regulator Arpin Identifies Its Acidic Tail as a Primary Binding Epitope.
Structure 24(2):252-60 (2016)
Fetics S, Thureau A, Campanacci V, Aumont-Nicaise M, Dang I, Gautreau A, Pérez J, Cherfils J
|
| RgGuinier |
1.8 |
nm |
| Dmax |
6.3 |
nm |
| VolumePorod |
24 |
nm3 |
|
|
UniProt ID: Q7Z6K5 (None-None) Human Arpin
|
|
|
|
| Sample: |
Human Arpin monomer, 25 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES 100mM NaCl 1mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2014 Dec 4
|
Hybrid Structural Analysis of the Arp2/3 Regulator Arpin Identifies Its Acidic Tail as a Primary Binding Epitope.
Structure 24(2):252-60 (2016)
Fetics S, Thureau A, Campanacci V, Aumont-Nicaise M, Dang I, Gautreau A, Pérez J, Cherfils J
|
| RgGuinier |
2.6 |
nm |
| Dmax |
13.2 |
nm |
| VolumePorod |
47 |
nm3 |
|
|
UniProt ID: Q1LWJ6 (None-None) Zebrafish (Danio rerio) Arpin
|
|
|
|
| Sample: |
Zebrafish (Danio rerio) Arpin monomer, 25 kDa Danio rerio protein
|
| Buffer: |
50 mM HEPES 100mM NaCl 1mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2014 Dec 4
|
Hybrid Structural Analysis of the Arp2/3 Regulator Arpin Identifies Its Acidic Tail as a Primary Binding Epitope.
Structure 24(2):252-60 (2016)
Fetics S, Thureau A, Campanacci V, Aumont-Nicaise M, Dang I, Gautreau A, Pérez J, Cherfils J
|
| RgGuinier |
2.7 |
nm |
| Dmax |
13.8 |
nm |
| VolumePorod |
47 |
nm3 |
|
|
UniProt ID: Q1LWJ6 (1-210) Zebrafish (Danio rerio) Arpin truncated C-terminal mutant (delta-C 16).
|
|
|
|
| Sample: |
Zebrafish (Danio rerio) Arpin truncated C-terminal mutant (delta-C 16). monomer, 24 kDa Danio rerio protein
|
| Buffer: |
50 mM HEPES 100mM NaCl 1mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2014 Dec 4
|
Hybrid Structural Analysis of the Arp2/3 Regulator Arpin Identifies Its Acidic Tail as a Primary Binding Epitope.
Structure 24(2):252-60 (2016)
Fetics S, Thureau A, Campanacci V, Aumont-Nicaise M, Dang I, Gautreau A, Pérez J, Cherfils J
|
| RgGuinier |
2.2 |
nm |
| Dmax |
11.3 |
nm |
| VolumePorod |
37 |
nm3 |
|
|
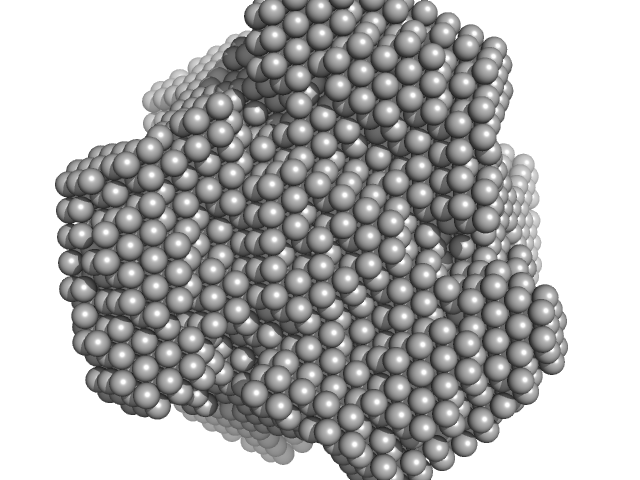
UniProt ID: P0A7A9 (None-None) Inorganic pyrophosphatase (PPase) from E. coli
|
|
|
|
| Sample: |
Inorganic pyrophosphatase (PPase) from E. coli hexamer, 117 kDa Escherichia coli protein
|
| Buffer: |
50 mM Tris 10 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Jun 30
|
X-Ray Solution Scattering Study of Four Escherichia coli Enzymes Involved in Stationary-Phase Metabolism.
PLoS One 11(5):e0156105 (2016)
Dadinova LA, Shtykova EV, Konarev PV, Rodina EV, Snalina NE, Vorobyeva NN, Kurilova SA, Nazarova TI, Jeffries CM, Svergun DI
|
| RgGuinier |
3.0 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
166 |
nm3 |
|
|
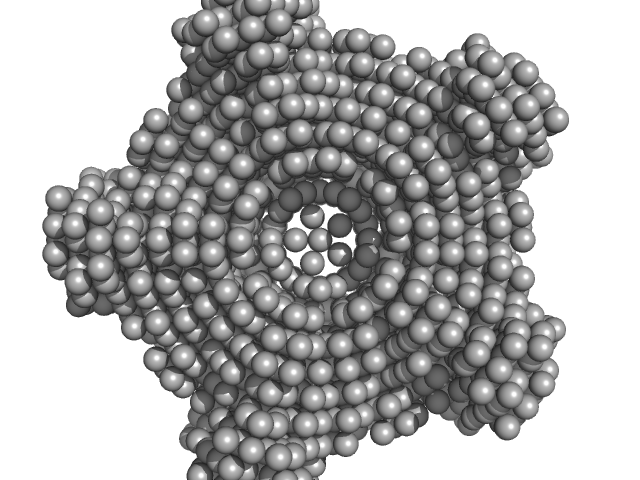
UniProt ID: A0A0K4BP99 (None-None) Class I fructose-1,6-bisphosphate aldolase (FbaB) from E. coli
|
|
|
|
| Sample: |
Class I fructose-1,6-bisphosphate aldolase (FbaB) from E. coli decamer, 381 kDa Escherichia coli protein
|
| Buffer: |
50 mM Tris 10 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Jun 30
|
X-Ray Solution Scattering Study of Four Escherichia coli Enzymes Involved in Stationary-Phase Metabolism.
PLoS One 11(5):e0156105 (2016)
Dadinova LA, Shtykova EV, Konarev PV, Rodina EV, Snalina NE, Vorobyeva NN, Kurilova SA, Nazarova TI, Jeffries CM, Svergun DI
|
| RgGuinier |
4.4 |
nm |
| Dmax |
12.7 |
nm |
| VolumePorod |
484 |
nm3 |
|
|
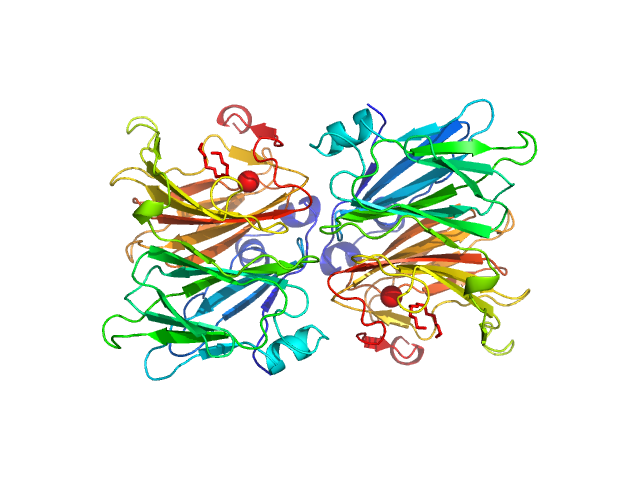
UniProt ID: Q46938 (None-None) 5-keto-4-deoxyuronate isomerase (KduI) from E. coli
|
|
|
|
| Sample: |
5-keto-4-deoxyuronate isomerase (KduI) from E. coli None, Escherichia coli protein
|
| Buffer: |
50 mM Tris 10 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Jun 20
|
X-Ray Solution Scattering Study of Four Escherichia coli Enzymes Involved in Stationary-Phase Metabolism.
PLoS One 11(5):e0156105 (2016)
Dadinova LA, Shtykova EV, Konarev PV, Rodina EV, Snalina NE, Vorobyeva NN, Kurilova SA, Nazarova TI, Jeffries CM, Svergun DI
|
|
|
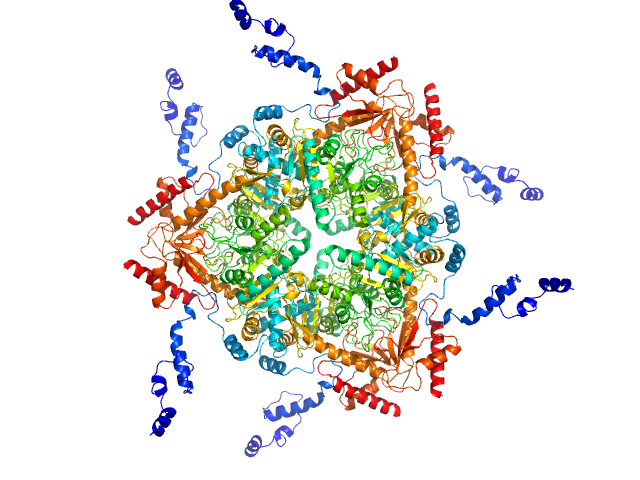
UniProt ID: P69908 (None-None) Glutamate decarboxylase alpha (GadA) from E. coli
|
|
|
|
| Sample: |
Glutamate decarboxylase alpha (GadA) from E. coli monomer, 53 kDa Escherichia coli protein
|
| Buffer: |
50 mM Tris 10 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Jun 20
|
X-Ray Solution Scattering Study of Four Escherichia coli Enzymes Involved in Stationary-Phase Metabolism.
PLoS One 11(5):e0156105 (2016)
Dadinova LA, Shtykova EV, Konarev PV, Rodina EV, Snalina NE, Vorobyeva NN, Kurilova SA, Nazarova TI, Jeffries CM, Svergun DI
|
| RgGuinier |
4.8 |
nm |
| VolumePorod |
410 |
nm3 |
|
|