UniProt ID: P11532 (718-1368) Dystrophin central domain repeats 4 to 9
|
|
|

|
| Sample: |
Dystrophin central domain repeats 4 to 9 monomer, 76 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris 150 mM NaCl 1 mM EDTA 2% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2012 Sep 19
|
Dystrophin's central domain forms a complex filament that becomes disorganized by in-frame deletions.
J Biol Chem 293(18):6637-6646 (2018)
Delalande O, Molza AE, Dos Santos Morais R, Chéron A, Pollet É, Raguenes-Nicol C, Tascon C, Giudice E, Guilbaud M, Nicolas A, Bondon A, Leturcq F, Férey N, Baaden M, Perez J, Roblin P, Piétri-Rouxel F, Hubert JF, Czjzek M, Le Rumeur E
|
| RgGuinier |
7.7 |
nm |
| Dmax |
30.5 |
nm |
| VolumePorod |
123 |
nm3 |
|
|
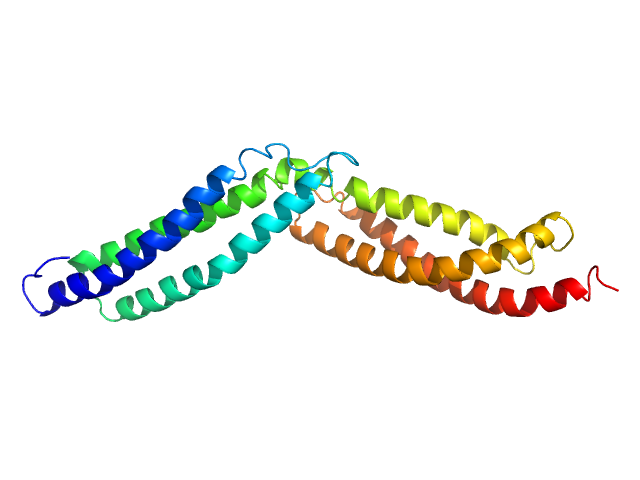
UniProt ID: P11532 (1984-2216) Dystrophin central domain repeats 16 to 17.
|
|
|
|
| Sample: |
Dystrophin central domain repeats 16 to 17. monomer, 28 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris 150 mM NaCl 1 mM EDTA 2% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2012 Sep 19
|
Dystrophin's central domain forms a complex filament that becomes disorganized by in-frame deletions.
J Biol Chem 293(18):6637-6646 (2018)
Delalande O, Molza AE, Dos Santos Morais R, Chéron A, Pollet É, Raguenes-Nicol C, Tascon C, Giudice E, Guilbaud M, Nicolas A, Bondon A, Leturcq F, Férey N, Baaden M, Perez J, Roblin P, Piétri-Rouxel F, Hubert JF, Czjzek M, Le Rumeur E
|
| RgGuinier |
3.1 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
47 |
nm3 |
|
|
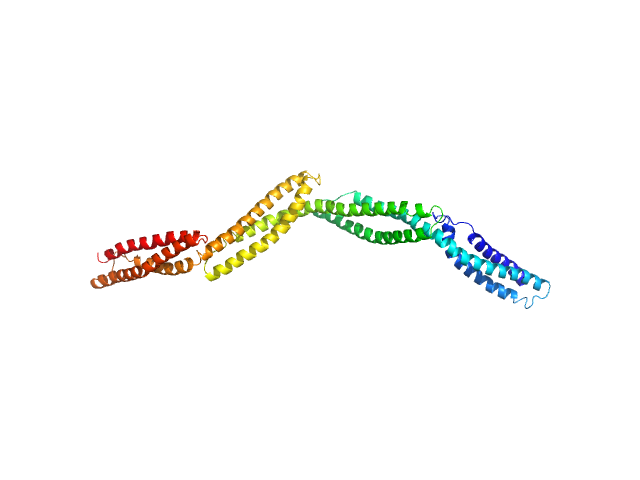
UniProt ID: P11532 (1994-2420) Dystrophin central domain repeats 16 to 19.
|
|
|
|
| Sample: |
Dystrophin central domain repeats 16 to 19. monomer, 50 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris 150 mM NaCl 1 mM EDTA 2% glycerol 5% acetonitrile, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2013 Oct 4
|
Dystrophin's central domain forms a complex filament that becomes disorganized by in-frame deletions.
J Biol Chem 293(18):6637-6646 (2018)
Delalande O, Molza AE, Dos Santos Morais R, Chéron A, Pollet É, Raguenes-Nicol C, Tascon C, Giudice E, Guilbaud M, Nicolas A, Bondon A, Leturcq F, Férey N, Baaden M, Perez J, Roblin P, Piétri-Rouxel F, Hubert JF, Czjzek M, Le Rumeur E
|
| RgGuinier |
4.6 |
nm |
| Dmax |
20.0 |
nm |
| VolumePorod |
70 |
nm3 |
|
|
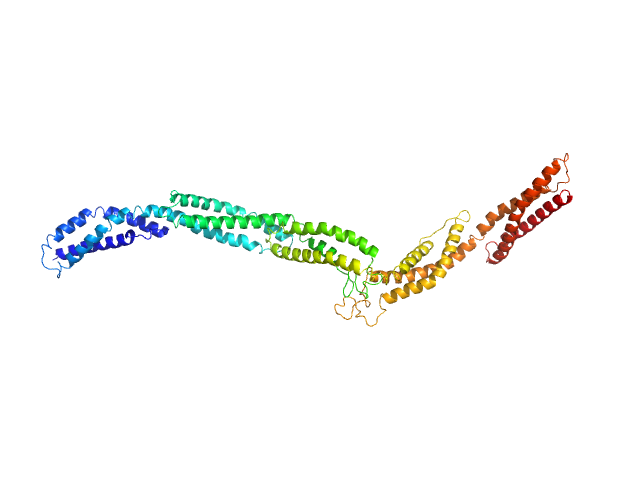
UniProt ID: P11532 (2469-3040) Dystrophin central domain repeats 20 to 24.
|
|
|
|
| Sample: |
Dystrophin central domain repeats 20 to 24. monomer, 67 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris 150 mM NaCl 1 mM EDTA 2% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2011 Jul 10
|
Dystrophin's central domain forms a complex filament that becomes disorganized by in-frame deletions.
J Biol Chem 293(18):6637-6646 (2018)
Delalande O, Molza AE, Dos Santos Morais R, Chéron A, Pollet É, Raguenes-Nicol C, Tascon C, Giudice E, Guilbaud M, Nicolas A, Bondon A, Leturcq F, Férey N, Baaden M, Perez J, Roblin P, Piétri-Rouxel F, Hubert JF, Czjzek M, Le Rumeur E
|
| RgGuinier |
5.8 |
nm |
| Dmax |
22.5 |
nm |
| VolumePorod |
107 |
nm3 |
|
|
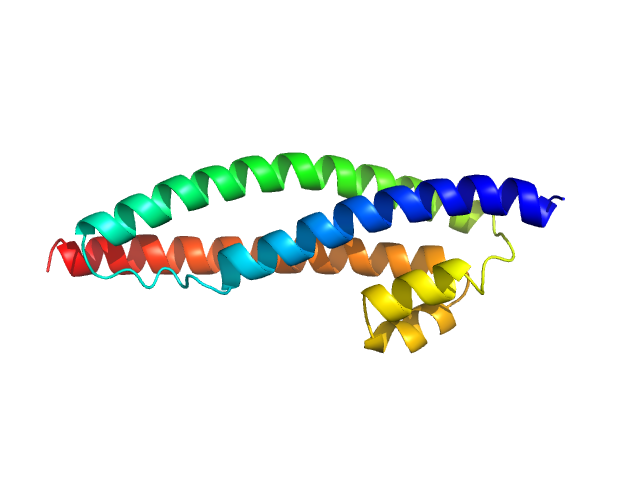
UniProt ID: P11532 (2800-2939) Dystrophin central domain single repeat 23
|
|
|
|
| Sample: |
Dystrophin central domain single repeat 23 monomer, 17 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris 150 mM NaCl 1 mM EDTA 2% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2011 Oct 7
|
Dystrophin's central domain forms a complex filament that becomes disorganized by in-frame deletions.
J Biol Chem 293(18):6637-6646 (2018)
Delalande O, Molza AE, Dos Santos Morais R, Chéron A, Pollet É, Raguenes-Nicol C, Tascon C, Giudice E, Guilbaud M, Nicolas A, Bondon A, Leturcq F, Férey N, Baaden M, Perez J, Roblin P, Piétri-Rouxel F, Hubert JF, Czjzek M, Le Rumeur E
|
| RgGuinier |
2.2 |
nm |
| Dmax |
7.4 |
nm |
| VolumePorod |
20 |
nm3 |
|
|
UniProt ID: Q8BGA3 (30-380) Mouse Leucine-rich repeat transmembrane neuronal protein 2, cLRRTM2 (stability engineered construct)
|
|
|

|
| Sample: |
Mouse Leucine-rich repeat transmembrane neuronal protein 2, cLRRTM2 (stability engineered construct) monomer, 40 kDa Mus musculus protein
|
| Buffer: |
20 mM Tris 150 mM NaCl 3% glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at ID14-3, ESRF on 2015 Sep 27
|
Crystal Structure of an Engineered LRRTM2 Synaptic Adhesion Molecule and a Model for Neurexin Binding.
Biochemistry 55(6):914-26 (2016)
Paatero A, Rosti K, Shkumatov AV, Sele C, Brunello C, Kysenius K, Singha P, Jokinen V, Huttunen H, Kajander T
|
| RgGuinier |
3.3 |
nm |
| Dmax |
13.1 |
nm |
|
|
UniProt ID: Q8BGA3 (30-380) Mouse Leucine-rich repeat transmembrane neuronal protein 2, LRRTM2
|
|
|

|
| Sample: |
Mouse Leucine-rich repeat transmembrane neuronal protein 2, LRRTM2 dimer, 80 kDa Mus musculus protein
|
| Buffer: |
20 mM Tris 150 mM NaCl 3% glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at ID14-3, ESRF on 2015 Jun 28
|
Crystal Structure of an Engineered LRRTM2 Synaptic Adhesion Molecule and a Model for Neurexin Binding.
Biochemistry 55(6):914-26 (2016)
Paatero A, Rosti K, Shkumatov AV, Sele C, Brunello C, Kysenius K, Singha P, Jokinen V, Huttunen H, Kajander T
|
| RgGuinier |
4.2 |
nm |
| Dmax |
21.6 |
nm |
|
|
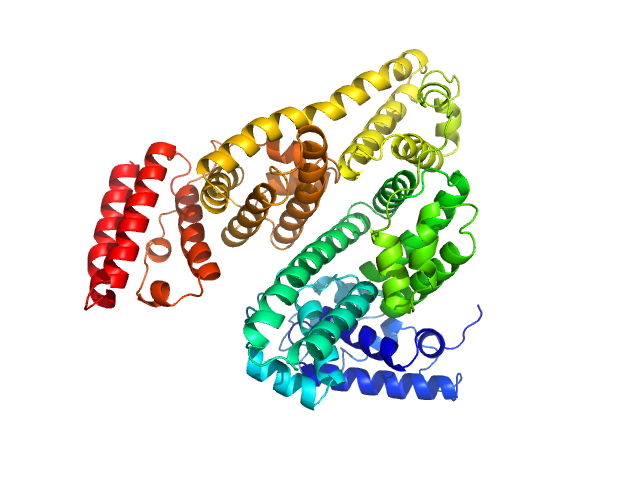
UniProt ID: P02769 (25-607) Bovine serum albumin, monomer
|
|
|
|
| Sample: |
Bovine serum albumin, monomer monomer, 66 kDa Bos taurus protein
|
| Buffer: |
25 mM Tris 150 mM NaCl 3% (v/v) glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2014 Jan 23
|
Preparing monodisperse macromolecular samples for successful biological small-angle X-ray and neutron-scattering experiments.
Nat Protoc 11(11):2122-2153 (2016)
Jeffries CM, Graewert MA, Blanchet CE, Langley DB, Whitten AE, Svergun DI
|
| RgGuinier |
2.8 |
nm |
| Dmax |
8.2 |
nm |
| VolumePorod |
100 |
nm3 |
|
|
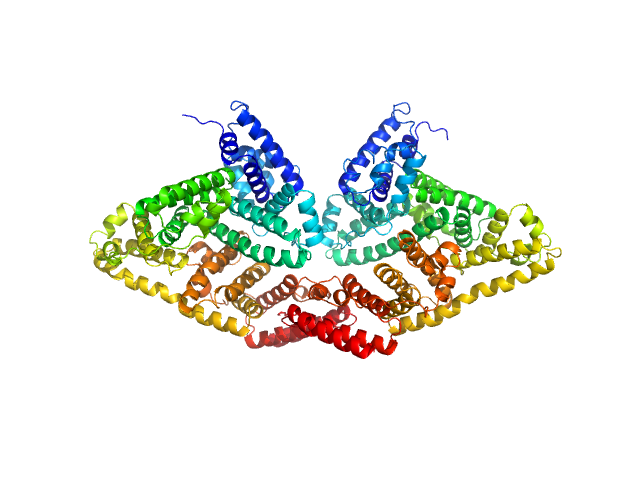
UniProt ID: P02769 (25-607) Bovine serum albumin, dimer
|
|
|
|
| Sample: |
Bovine serum albumin, dimer dimer, 133 kDa Bos taurus protein
|
| Buffer: |
25 mM Tris 150 mM NaCl 3% (v/v) glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2014 Jan 23
|
Preparing monodisperse macromolecular samples for successful biological small-angle X-ray and neutron-scattering experiments.
Nat Protoc 11(11):2122-2153 (2016)
Jeffries CM, Graewert MA, Blanchet CE, Langley DB, Whitten AE, Svergun DI
|
| RgGuinier |
3.9 |
nm |
| Dmax |
12.7 |
nm |
| VolumePorod |
202 |
nm3 |
|
|
UniProt ID: Q6ZSD7 (1-400) cDNA FLJ45612 fis, clone BRTHA3025073, highly similar to Actin cross-linking family protein 7
|
|
|
|
| Sample: |
CDNA FLJ45612 fis, clone BRTHA3025073, highly similar to Actin cross-linking family protein 7 monomer, 46 kDa Homo sapiens protein
|
| Buffer: |
PBS, pH: 7.4 |
| Experiment: |
SAXS
data collected at 12-ID-B SAXS/WAXS, Advanced Photon Source (APS), Argonne National Laboratory on 2014 Nov 10
|
In vivo epidermal migration requires focal adhesion targeting of ACF7.
Nat Commun 7:11692 (2016)
Yue J, Zhang Y, Liang WG, Gou X, Lee P, Liu H, Lyu W, Tang WJ, Chen SY, Yang F, Liang H, Wu X
|
| RgGuinier |
3.4 |
nm |
| Dmax |
13.5 |
nm |
| VolumePorod |
53 |
nm3 |
|
|