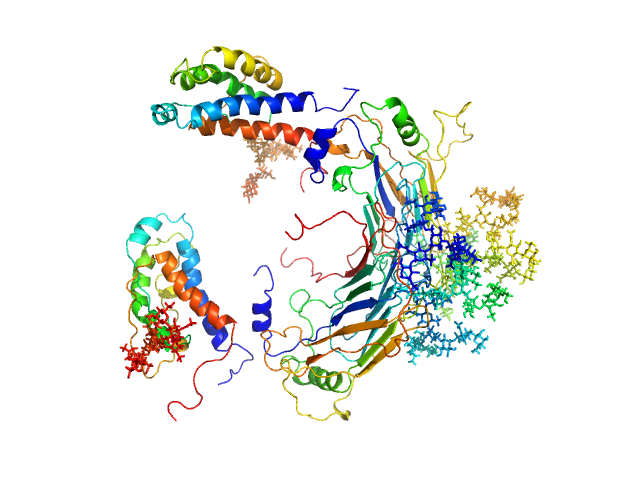
UniProt ID: Q9J5U5 (None-None) GM-CSF/IL-2 inhibition factor
UniProt ID: P19114 (21-155) Interleukin-2
|
|
|
|
| Sample: |
GM-CSF/IL-2 inhibition factor tetramer, 120 kDa Orf virus protein
Interleukin-2 monomer, 16 kDa Ovis aries protein
|
| Buffer: |
20 mM HEPES 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2014 Sep 10
|
Structural basis of GM-CSF and IL-2 sequestration by the viral decoy receptor GIF.
Nat Commun 7:13228 (2016)
Felix J, Kandiah E, De Munck S, Bloch Y, van Zundert GC, Pauwels K, Dansercoer A, Novanska K, Read RJ, Bonvin AM, Vergauwen B, Verstraete K, Gutsche I, Savvides SN
|
| RgGuinier |
4.1 |
nm |
| Dmax |
12.9 |
nm |
| VolumePorod |
253 |
nm3 |
|
|
UniProt ID: P21333 (1782-1956) Human Filamin A Ig-like domains 16-17
|
|
|

|
| Sample: |
Human Filamin A Ig-like domains 16-17 monomer, 19 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris 50 mM NaCl 10 mM DTT, pH: |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2013 Sep 30
|
Skeletal Dysplasia Mutations Effect on Human Filamins' Structure and Mechanosensing.
Sci Rep 7(1):4218 (2017)
Seppälä J, Bernardi RC, Haataja TJK, Hellman M, Pentikäinen OT, Schulten K, Permi P, Ylänne J, Pentikäinen U
|
| RgGuinier |
1.8 |
nm |
| Dmax |
6.2 |
nm |
| VolumePorod |
31 |
nm3 |
|
|
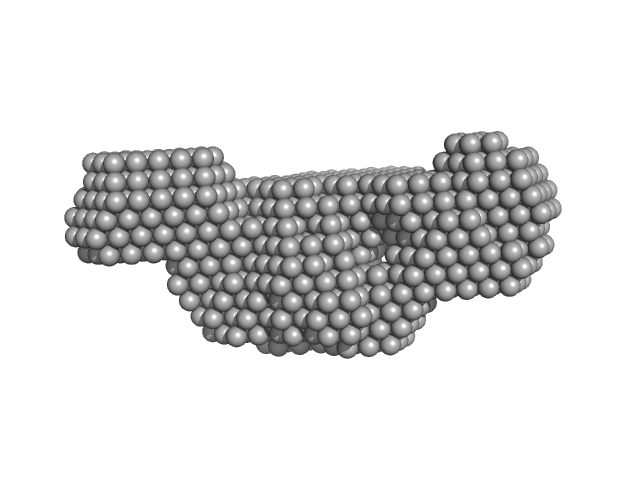
UniProt ID: P21333 (1781-1956) Human Filamin A Ig-like domains 16-17*
|
|
|
|
| Sample: |
Human Filamin A Ig-like domains 16-17* monomer, 19 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris 50 mM NaCl 10 mM DTT, pH: |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2013 Sep 30
|
Skeletal Dysplasia Mutations Effect on Human Filamins' Structure and Mechanosensing.
Sci Rep 7(1):4218 (2017)
Seppälä J, Bernardi RC, Haataja TJK, Hellman M, Pentikäinen OT, Schulten K, Permi P, Ylänne J, Pentikäinen U
|
| RgGuinier |
2.4 |
nm |
| Dmax |
8.3 |
nm |
| VolumePorod |
37 |
nm3 |
|
|
UniProt ID: Q4JBH1 (1-116) Wild-type archaeal biofilm regulator 1 (ABfR1: Transcriptional regulator ArsR family).
|
|
|

|
| Sample: |
Wild-type archaeal biofilm regulator 1 (ABfR1: Transcriptional regulator ArsR family). dimer, 26 kDa Sulfolobus acidocaldarius protein
|
| Buffer: |
20 mM HEPES 200 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2015 Aug 27
|
Wing phosphorylation is a major functional determinant of the Lrs14-type biofilm and motility regulator AbfR1 in Sulfolobus acidocaldarius.
Mol Microbiol 105(5):777-793 (2017)
Li L, Banerjee A, Bischof LF, Maklad HR, Hoffmann L, Henche AL, Veliz F, Bildl W, Schulte U, Orell A, Essen LO, Peeters E, Albers SV
|
| RgGuinier |
2.6 |
nm |
| Dmax |
9.8 |
nm |
| VolumePorod |
47 |
nm3 |
|
|
UniProt ID: Q4JBH1 (1-116) Archaeal biofilm regulator 1 (AbfR1) mutant Y84E S87D phosphomimic mutant
|
|
|

|
| Sample: |
Archaeal biofilm regulator 1 (AbfR1) mutant Y84E S87D phosphomimic mutant dimer, 26 kDa Sulfolobus acidocaldarius protein
|
| Buffer: |
20 mM HEPES 200 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2015 Aug 27
|
Wing phosphorylation is a major functional determinant of the Lrs14-type biofilm and motility regulator AbfR1 in Sulfolobus acidocaldarius.
Mol Microbiol 105(5):777-793 (2017)
Li L, Banerjee A, Bischof LF, Maklad HR, Hoffmann L, Henche AL, Veliz F, Bildl W, Schulte U, Orell A, Essen LO, Peeters E, Albers SV
|
| RgGuinier |
2.6 |
nm |
| Dmax |
9.5 |
nm |
| VolumePorod |
51 |
nm3 |
|
|
UniProt ID: (None-None) 40bp long dsDNA-Sa Oligonucleotide
UniProt ID: Q4JBH1 (1-116) Wild-type archaeal biofilm regulator 1 (ABfR1: Transcriptional regulator ArsR family).
|
|
|

|
| Sample: |
40bp long dsDNA-Sa Oligonucleotide monomer, 25 kDa DNA
Wild-type archaeal biofilm regulator 1 (ABfR1: Transcriptional regulator ArsR family). dimer, 26 kDa Sulfolobus acidocaldarius protein
|
| Buffer: |
0.5 x Tris/Borate/EDTA (TBE), pH: |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2015 Aug 27
|
Wing phosphorylation is a major functional determinant of the Lrs14-type biofilm and motility regulator AbfR1 in Sulfolobus acidocaldarius.
Mol Microbiol 105(5):777-793 (2017)
Li L, Banerjee A, Bischof LF, Maklad HR, Hoffmann L, Henche AL, Veliz F, Bildl W, Schulte U, Orell A, Essen LO, Peeters E, Albers SV
|
| RgGuinier |
3.0 |
nm |
| Dmax |
11.8 |
nm |
|
|
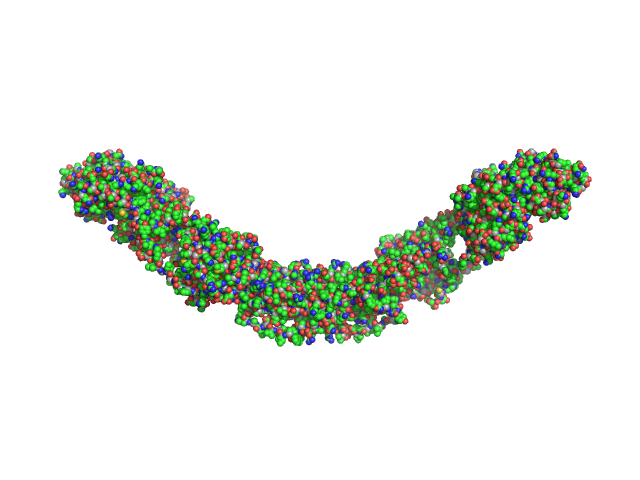
UniProt ID: Q9NRD5 (None-None) Maltose Binding Protein fused to Protein Interacting with C kinase 1
|
|
|
|
| Sample: |
Maltose Binding Protein fused to Protein Interacting with C kinase 1 dimer, 174 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris 300 mM NaCl 1 mM maltose 1 mM EGTA 2 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2015 Oct 13
|
PICK1 is implicated in organelle motility in an Arp2/3 complex-independent manner.
Mol Biol Cell 26(7):1308-22 (2015)
Madasu Y, Yang C, Boczkowska M, Bethoney KA, Zwolak A, Rebowski G, Svitkina T, Dominguez R
|
| RgGuinier |
8.4 |
nm |
| Dmax |
27.6 |
nm |
| VolumePorod |
483 |
nm3 |
|
|
UniProt ID: Q5U5U6 (1-152) Linear di-ubiquitin
|
|
|
|
| Sample: |
Linear di-ubiquitin monomer, 17 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris 150 mM NaCl 1 mM MgCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at 5C, Pohang Accelerator Laboratory on 2014 Nov 3
|
New conformations of linear polyubiquitin chains from crystallographic and solution-scattering studies expand the conformational space of polyubiquitin.
Acta Crystallogr D Struct Biol 72(Pt 4):524-35 (2016)
Thach TT, Shin D, Han S, Lee S
|
| RgGuinier |
2.1 |
nm |
| Dmax |
6.6 |
nm |
| VolumePorod |
20 |
nm3 |
|
|
UniProt ID: Q5U5U6 (None-None) Human linear tri-ubiquitin
|
|
|
|
| Sample: |
Human linear tri-ubiquitin monomer, 26 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris 150mM NaCl 0.5 mM EDTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at 5C, Pohang Accelerator Laboratory on 2014 Nov 3
|
New conformations of linear polyubiquitin chains from crystallographic and solution-scattering studies expand the conformational space of polyubiquitin.
Acta Crystallogr D Struct Biol 72(Pt 4):524-35 (2016)
Thach TT, Shin D, Han S, Lee S
|
| RgGuinier |
2.5 |
nm |
| Dmax |
8.6 |
nm |
| VolumePorod |
36 |
nm3 |
|
|
UniProt ID: Q5U5U6 (None-None) Human linear tetra-ubiquitin
|
|
|
|
| Sample: |
Human linear tetra-ubiquitin monomer, 34 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris 150mM NaCl 0.5 mM EDTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at 5C, Pohang Accelerator Laboratory on 2014 Nov 3
|
New conformations of linear polyubiquitin chains from crystallographic and solution-scattering studies expand the conformational space of polyubiquitin.
Acta Crystallogr D Struct Biol 72(Pt 4):524-35 (2016)
Thach TT, Shin D, Han S, Lee S
|
| RgGuinier |
3.1 |
nm |
| Dmax |
11.2 |
nm |
| VolumePorod |
49 |
nm3 |
|
|