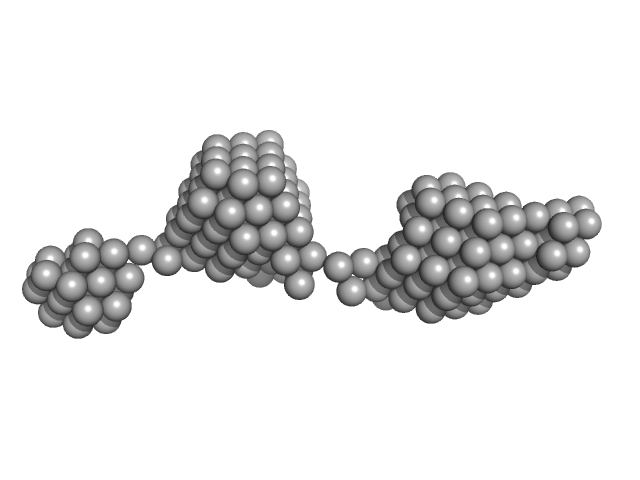
UniProt ID: Q0B303 (2616-2644) Beta-ketoacyl synthase Bamb_5925
|
|
|
|
| Sample: |
Beta-ketoacyl synthase Bamb_5925 monomer, 4 kDa Burkholderia ambifaria protein
|
| Buffer: |
20 mM Tris-HCl, 200 mM NaCl, 5% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Jan 23
|
Towards improved understanding of intersubunit interactions in modular polyketide biosynthesis: docking in the enacyloxin IIa polyketide synthase.
J Struct Biol :107581 (2020)
Risser F, Collin S, Dos Santos-Morais R, Gruez A, Chagot B, Weissman KJ
|
| RgGuinier |
1.6 |
nm |
| Dmax |
6.2 |
nm |
| VolumePorod |
3 |
nm3 |
|
|
UniProt ID: Q0B304 (1-41) Beta-ketoacyl synthase Bamb_5924
|
|
|

|
| Sample: |
Beta-ketoacyl synthase Bamb_5924 dimer, 10 kDa Burkholderia ambifaria protein
|
| Buffer: |
20 mM Tris-HCl, 200 mM NaCl, 5% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2017 Oct 6
|
Towards improved understanding of intersubunit interactions in modular polyketide biosynthesis: docking in the enacyloxin IIa polyketide synthase.
J Struct Biol :107581 (2020)
Risser F, Collin S, Dos Santos-Morais R, Gruez A, Chagot B, Weissman KJ
|
| RgGuinier |
1.9 |
nm |
| Dmax |
7.5 |
nm |
| VolumePorod |
22 |
nm3 |
|
|
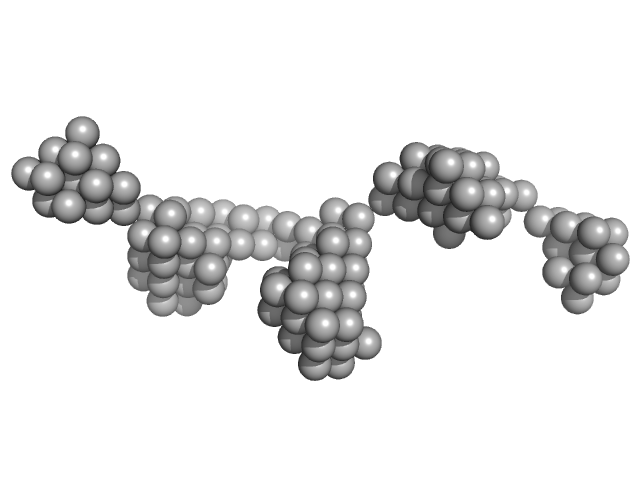
UniProt ID: Q0B308 (2956-2975) Beta-ketoacyl synthase Bamb_5920
|
|
|
|
| Sample: |
Beta-ketoacyl synthase Bamb_5920 monomer, 2 kDa Burkholderia ambifaria protein
|
| Buffer: |
20 mM Tris-HCl, 200 mM NaCl, 5% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Jan 23
|
Towards improved understanding of intersubunit interactions in modular polyketide biosynthesis: docking in the enacyloxin IIa polyketide synthase.
J Struct Biol :107581 (2020)
Risser F, Collin S, Dos Santos-Morais R, Gruez A, Chagot B, Weissman KJ
|
| RgGuinier |
1.2 |
nm |
| Dmax |
4.9 |
nm |
| VolumePorod |
3 |
nm3 |
|
|
UniProt ID: Q0B304 (1-41) Beta-ketoacyl synthase Bamb_5924, K32A mutant
|
|
|
|
| Sample: |
Beta-ketoacyl synthase Bamb_5924, K32A mutant dimer, 10 kDa Burkholderia ambifaria protein
|
| Buffer: |
20 mM Tris-HCl, 200 mM NaCl, 5% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Jan 23
|
Towards improved understanding of intersubunit interactions in modular polyketide biosynthesis: docking in the enacyloxin IIa polyketide synthase.
J Struct Biol :107581 (2020)
Risser F, Collin S, Dos Santos-Morais R, Gruez A, Chagot B, Weissman KJ
|
| RgGuinier |
1.8 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
22 |
nm3 |
|
|
UniProt ID: Q0B304 (1-41) Beta-ketoacyl synthase Bamb_5924, R17A mutant
|
|
|
|
| Sample: |
Beta-ketoacyl synthase Bamb_5924, R17A mutant dimer, 10 kDa Burkholderia ambifaria protein
|
| Buffer: |
20 mM Tris-HCl, 200 mM NaCl, 5% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Jan 23
|
Towards improved understanding of intersubunit interactions in modular polyketide biosynthesis: docking in the enacyloxin IIa polyketide synthase.
J Struct Biol :107581 (2020)
Risser F, Collin S, Dos Santos-Morais R, Gruez A, Chagot B, Weissman KJ
|
| RgGuinier |
1.8 |
nm |
| Dmax |
8.2 |
nm |
| VolumePorod |
20 |
nm3 |
|
|
UniProt ID: M9NUS9 (1-381) Major tail protein
|
|
|

|
| Sample: |
Major tail protein monomer, 40 kDa Salmonella virus Chi protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 0.03 % NaN3, 5.0 % glycerol, pH: 7.8 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2017 Mar 8
|
The architecture and stabilisation of flagellotropic tailed bacteriophages.
Nat Commun 11(1):3748 (2020)
Hardy JM, Dunstan RA, Grinter R, Belousoff MJ, Wang J, Pickard D, Venugopal H, Dougan G, Lithgow T, Coulibaly F
|
| RgGuinier |
3.3 |
nm |
| Dmax |
10.2 |
nm |
| VolumePorod |
54 |
nm3 |
|
|
UniProt ID: P13433 (30-1351) DNA-directed RNA polymerase
|
|
|
|
| Sample: |
DNA-directed RNA polymerase monomer, 150 kDa Saccharomyces cerevisiae protein
|
| Buffer: |
40 mM Tris, 150 mM NaCl, 5% glycerol, 1 mM DDT, 1 mM EDTA, pH: 8 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpace, CSIR-Central Drug Research Institute on 2019 Sep 25
|
PfKsgA1 functions as a transcription initiation factor and interacts with the N-terminal region of the mitochondrial RNA polymerase of Plasmodium falciparum.
Int J Parasitol (2020)
Gupta A, Shrivastava D, Shakya AK, Gupta K, Pratap JV, Habib S
|
| RgGuinier |
4.4 |
nm |
| Dmax |
12.5 |
nm |
| VolumePorod |
295 |
nm3 |
|
|
UniProt ID: P13433 (30-1351) DNA-directed RNA polymerase
UniProt ID: Q8ILT8 (43-381) rRNA adenine N (6) methyl transferase
|
|
|
|
| Sample: |
DNA-directed RNA polymerase monomer, 150 kDa Saccharomyces cerevisiae protein
RRNA adenine N (6) methyl transferase monomer, 39 kDa Plasmodium falciparum protein
|
| Buffer: |
40 mM Tris, 150 mM NaCl, 5% glycerol, 1 mM DDT, 1 mM EDTA, pH: 8 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpace, CSIR-Central Drug Research Institute on 2020 Jan 10
|
PfKsgA1 functions as a transcription initiation factor and interacts with the N-terminal region of the mitochondrial RNA polymerase of Plasmodium falciparum.
Int J Parasitol (2020)
Gupta A, Shrivastava D, Shakya AK, Gupta K, Pratap JV, Habib S
|
| RgGuinier |
5.5 |
nm |
| Dmax |
15.0 |
nm |
| VolumePorod |
393 |
nm3 |
|
|
UniProt ID: P13433 (30-1351) DNA-directed RNA polymerase
UniProt ID: P14908 (1-341) Mitochondrial transcription factor 1
|
|
|
|
| Sample: |
DNA-directed RNA polymerase monomer, 150 kDa Saccharomyces cerevisiae protein
Mitochondrial transcription factor 1 monomer, 40 kDa Saccharomyces cerevisiae protein
|
| Buffer: |
40 mM Tris, 150 mM NaCl, 5% glycerol, 1 mM DDT, 1 mM EDTA, pH: 8 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpace, CSIR-Central Drug Research Institute on 2019 Dec 7
|
PfKsgA1 functions as a transcription initiation factor and interacts with the N-terminal region of the mitochondrial RNA polymerase of Plasmodium falciparum.
Int J Parasitol (2020)
Gupta A, Shrivastava D, Shakya AK, Gupta K, Pratap JV, Habib S
|
| RgGuinier |
4.5 |
nm |
| Dmax |
14.4 |
nm |
| VolumePorod |
292 |
nm3 |
|
|
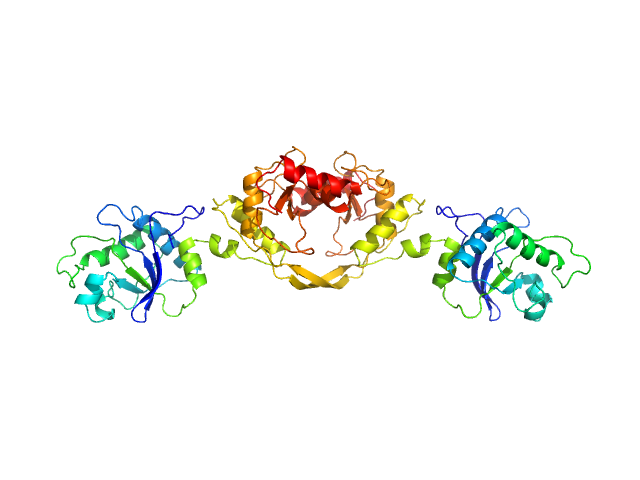
UniProt ID: P24200 (1-277) 5-methylcytosine-specific restriction enzyme A
|
|
|
|
| Sample: |
5-methylcytosine-specific restriction enzyme A dimer, 65 kDa Escherichia coli protein
|
| Buffer: |
20 mM Tris–HCl pH 7.5, 200 mM KCl, 0.1 mM EDTA, 0.01% (w/v) sodium azide and 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 May 24
|
Activity and structure of EcoKMcrA.
Nucleic Acids Res 46(18):9829-9841 (2018)
Czapinska H, Kowalska M, Zagorskaite E, Manakova E, Slyvka A, Xu SY, Siksnys V, Sasnauskas G, Bochtler M
|
| RgGuinier |
3.7 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
95 |
nm3 |
|
|