UniProt ID: G0SB82 (3-1222) Condensin complex subunit 1
UniProt ID: G0SBJ6 (225-418) Condensin complex subunit 2, 225-418
UniProt ID: G0SBJ6 (776-898) Condensin complex subunit 2, 776-898
UniProt ID: G0S2G2 (263-466) SMC hinge domain-containing protein, 263-466
UniProt ID: G0S2G2 (1367-1542) SMC hinge domain-containing protein, 1367-1542
|
|
|
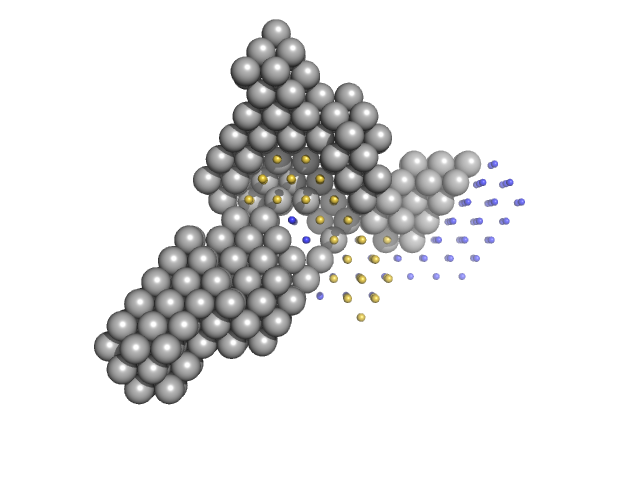
|
| Sample: |
Condensin complex subunit 1 monomer, 137 kDa Chaetomium thermophilum protein
Condensin complex subunit 2, 225-418 monomer, 21 kDa Chaetomium thermophilum protein
Condensin complex subunit 2, 776-898 monomer, 14 kDa Chaetomium thermophilum protein
SMC hinge domain-containing protein, 263-466 monomer, 22 kDa Chaetomium thermophilum protein
SMC hinge domain-containing protein, 1367-1542 monomer, 20 kDa Chaetomium thermophilum protein
|
| Buffer: |
25 mM Tris, 300 mM NaCl, 1mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Nov 20
|
Molecular flexibility of the condensin subunit Ycs4 is modulated by kleisin binding
Karen Manalastas-Cantos
|
| RgGuinier |
5.1 |
nm |
| Dmax |
17.9 |
nm |
| VolumePorod |
355 |
nm3 |
|
|
UniProt ID: P18887 (294-633) DNA repair protein XRCC1ΔN
|
|
|
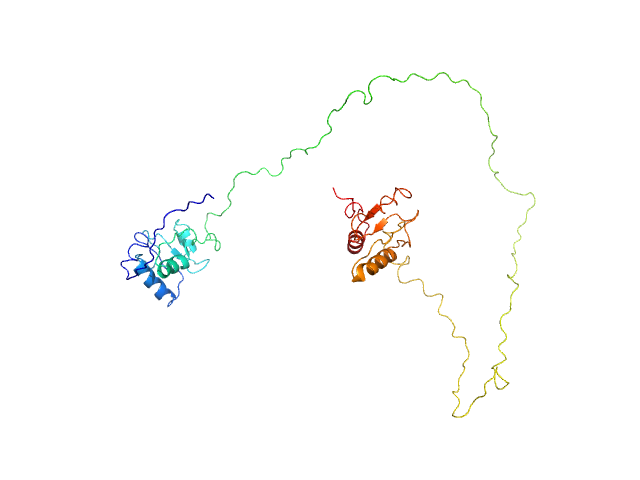
|
| Sample: |
DNA repair protein XRCC1ΔN dimer, 76 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris-HCl pH 7.5, 150 mM NaCl, 10% glycerol, 2 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2011 Jul 15
|
An atypical BRCT-BRCT interaction with the XRCC1 scaffold protein compacts human DNA Ligase IIIα within a flexible DNA repair complex.
Nucleic Acids Res (2020)
Hammel M, Rashid I, Sverzhinsky A, Pourfarjam Y, Tsai MS, Ellenberger T, Pascal JM, Kim IK, Tainer JA, Tomkinson AE
|
| RgGuinier |
5.4 |
nm |
| Dmax |
19.5 |
nm |
| VolumePorod |
170 |
nm3 |
|
|
UniProt ID: P18887 (1-633) DNA repair protein XRCC1
|
|
|
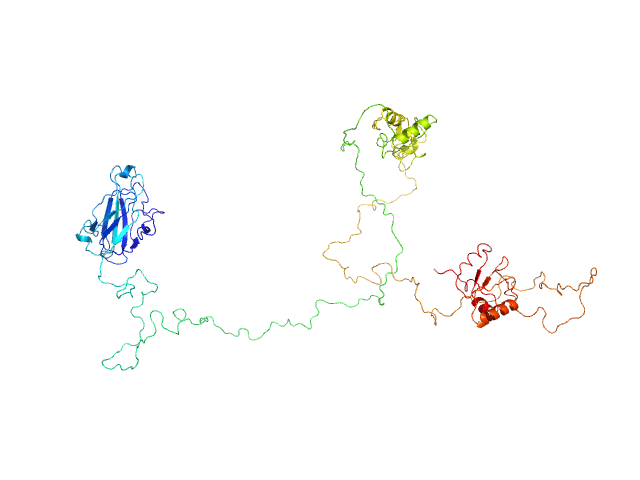
|
| Sample: |
DNA repair protein XRCC1, 69 kDa Homo sapiens protein
|
| Buffer: |
200 mM NaCl, 20 mM Tris-HCl, pH 7.5, 2% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 Jun 19
|
An atypical BRCT-BRCT interaction with the XRCC1 scaffold protein compacts human DNA Ligase IIIα within a flexible DNA repair complex.
Nucleic Acids Res (2020)
Hammel M, Rashid I, Sverzhinsky A, Pourfarjam Y, Tsai MS, Ellenberger T, Pascal JM, Kim IK, Tainer JA, Tomkinson AE
|
| RgGuinier |
6.3 |
nm |
| Dmax |
26.9 |
nm |
| VolumePorod |
330 |
nm3 |
|
|
UniProt ID: P49916 (88-1009) DNA ligase 3 (DNA ligase III alpha)
|
|
|
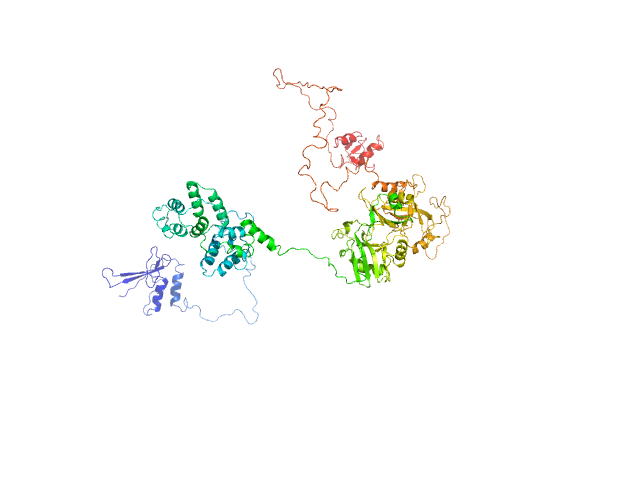
|
| Sample: |
DNA ligase 3 (DNA ligase III alpha), Homo sapiens protein
|
| Buffer: |
25 mM Tris-HCl pH 7.5, 150 mM NaCl, 10% glycerol, 2 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2011 Mar 31
|
An atypical BRCT-BRCT interaction with the XRCC1 scaffold protein compacts human DNA Ligase IIIα within a flexible DNA repair complex.
Nucleic Acids Res (2020)
Hammel M, Rashid I, Sverzhinsky A, Pourfarjam Y, Tsai MS, Ellenberger T, Pascal JM, Kim IK, Tainer JA, Tomkinson AE
|
| RgGuinier |
6.1 |
nm |
| Dmax |
21.0 |
nm |
| VolumePorod |
240 |
nm3 |
|
|
UniProt ID: P18887 (1-633) DNA repair protein XRCC1
UniProt ID: P49916 (88-1009) DNA ligase 3 (DNA ligase III alpha)
|
|
|
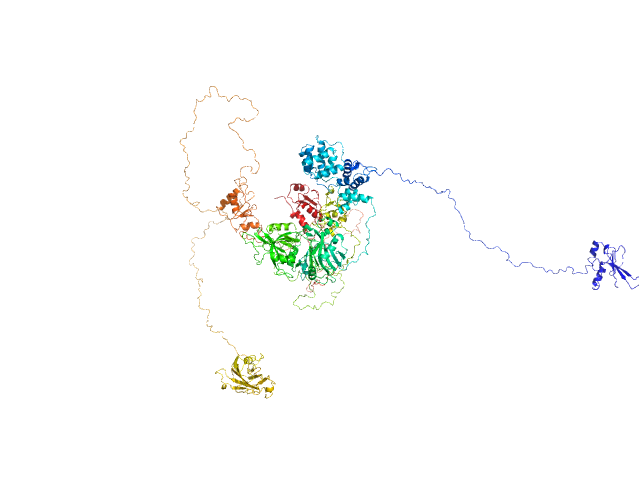
|
| Sample: |
DNA repair protein XRCC1, 69 kDa Homo sapiens protein
DNA ligase 3 (DNA ligase III alpha), Homo sapiens protein
|
| Buffer: |
50 mM Tris-HCl, pH 7.5, 100 mM NaCl, 5 mM MgCl₂, 0.2 mM PMSF, 1 mM benzamidine, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 Aug 18
|
An atypical BRCT-BRCT interaction with the XRCC1 scaffold protein compacts human DNA Ligase IIIα within a flexible DNA repair complex.
Nucleic Acids Res (2020)
Hammel M, Rashid I, Sverzhinsky A, Pourfarjam Y, Tsai MS, Ellenberger T, Pascal JM, Kim IK, Tainer JA, Tomkinson AE
|
| RgGuinier |
6.2 |
nm |
| Dmax |
27.9 |
nm |
| VolumePorod |
675 |
nm3 |
|
|
UniProt ID: Q5HPA2 (None-None) Extracellular matrix binding protein F-repeats
|
|
|
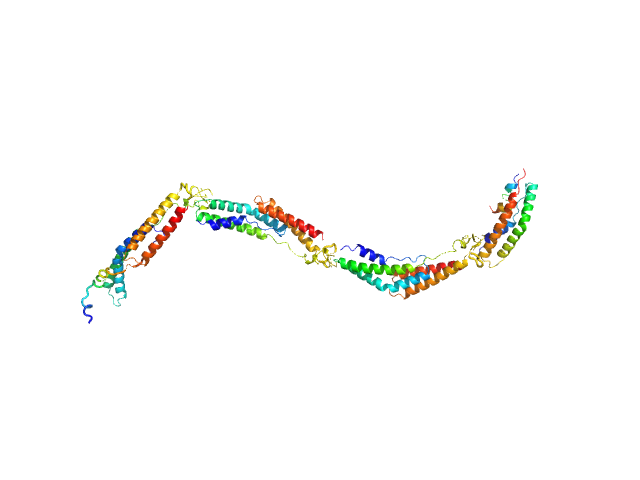
|
| Sample: |
Extracellular matrix binding protein F-repeats, 70 kDa Staphylococcus epidermidis protein
|
| Buffer: |
50 mM MES, 150 mM NaCl, pH: 6 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 Dec 15
|
A Giant Extracellular Matrix Binding Protein of Staphylococcus epidermidis Binds Surface-Immobilized Fibronectin via a Novel Mechanism.
mBio 11(5) (2020)
Büttner H, Perbandt M, Kohler T, Kikhney A, Wolters M, Christner M, Heise M, Wilde J, Weißelberg S, Both A, Betzel C, Hammerschmidt S, Svergun D, Aepfelbacher M, Rohde H
|
| RgGuinier |
7.0 |
nm |
| Dmax |
28.0 |
nm |
|
|
UniProt ID: Q5HPA2 (None-None) Extracellular matrix binding protein FG-repeats
|
|
|

|
| Sample: |
Extracellular matrix binding protein FG-repeats, 85 kDa Staphylococcus epidermidis protein
|
| Buffer: |
PBS, 136.5 mM NaCl, 2.65 mM KCl, 8.3 mM Na2HPO4, 2.65 mM KH2PO4, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Jun 14
|
A Giant Extracellular Matrix Binding Protein of Staphylococcus epidermidis Binds Surface-Immobilized Fibronectin via a Novel Mechanism.
mBio 11(5) (2020)
Büttner H, Perbandt M, Kohler T, Kikhney A, Wolters M, Christner M, Heise M, Wilde J, Weißelberg S, Both A, Betzel C, Hammerschmidt S, Svergun D, Aepfelbacher M, Rohde H
|
| RgGuinier |
10.7 |
nm |
| Dmax |
40.0 |
nm |
|
|
UniProt ID: Q9KHV6 (1-278) Type-2 restriction enzyme AgeI
|
|
|

|
| Sample: |
Type-2 restriction enzyme AgeI monomer, 31 kDa Thalassobius gelatinovorus protein
|
| Buffer: |
10 mM Tris-HCl, pH 7.5, 150 mM NaCl, 5 mM CaCl₂, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Nov 14
|
Restriction endonuclease AgeI is a monomer which dimerizes to cleave DNA.
Nucleic Acids Res 45(6):3547-3558 (2017)
Tamulaitiene G, Jovaisaite V, Tamulaitis G, Songailiene I, Manakova E, Zaremba M, Grazulis S, Xu SY, Siksnys V
|
| RgGuinier |
2.2 |
nm |
| Dmax |
6.7 |
nm |
| VolumePorod |
53 |
nm3 |
|
|
UniProt ID: Q9KHV6 (1-278) Type-2 restriction enzyme AgeI
UniProt ID: None (None-None) Cognate DNA oligoduplex with 5'-T overhang
|
|
|
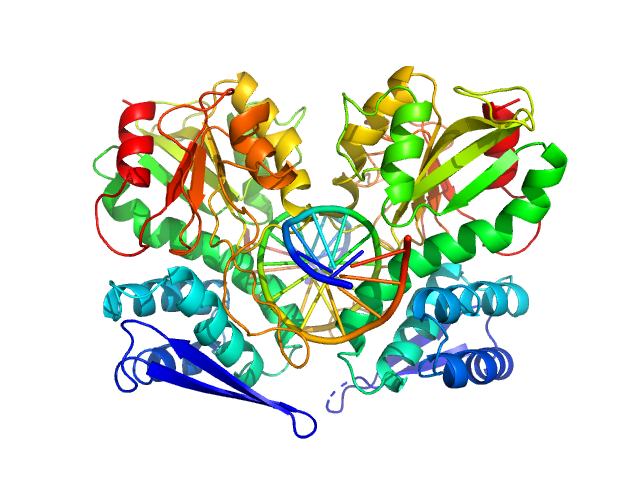
|
| Sample: |
Type-2 restriction enzyme AgeI dimer, 61 kDa Thalassobius gelatinovorus protein
Cognate DNA oligoduplex with 5'-T overhang dimer, 8 kDa DNA
|
| Buffer: |
10 mM Tris-HCl, pH 7.5, 150 mM NaCl, 5 mM CaCl₂, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Nov 14
|
Restriction endonuclease AgeI is a monomer which dimerizes to cleave DNA.
Nucleic Acids Res 45(6):3547-3558 (2017)
Tamulaitiene G, Jovaisaite V, Tamulaitis G, Songailiene I, Manakova E, Zaremba M, Grazulis S, Xu SY, Siksnys V
|
| RgGuinier |
2.4 |
nm |
| Dmax |
7.4 |
nm |
| VolumePorod |
69 |
nm3 |
|
|
UniProt ID: P12579 (127-163) Phosphoprotein
|
|
|
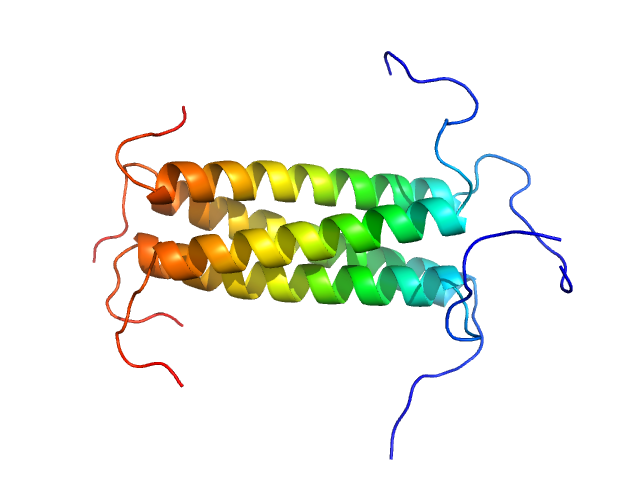
|
| Sample: |
Phosphoprotein tetramer, 18 kDa Respiratory syncytial virus protein
|
| Buffer: |
20 mM Na phosphate 100 mM NaCl, pH: 6.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Jun 21
|
A Structural and Dynamic Analysis of the Partially Disordered Polymerase-Binding Domain in RSV Phosphoprotein
Biomolecules 11(8):1225 (2021)
Cardone C, Caseau C, Bardiaux B, Thureaux A, Galloux M, Bajorek M, Eléouët J, Litaudon M, Bontems F, Sizun C
|
| RgGuinier |
2.1 |
nm |
| Dmax |
7.6 |
nm |
| VolumePorod |
33 |
nm3 |
|
|