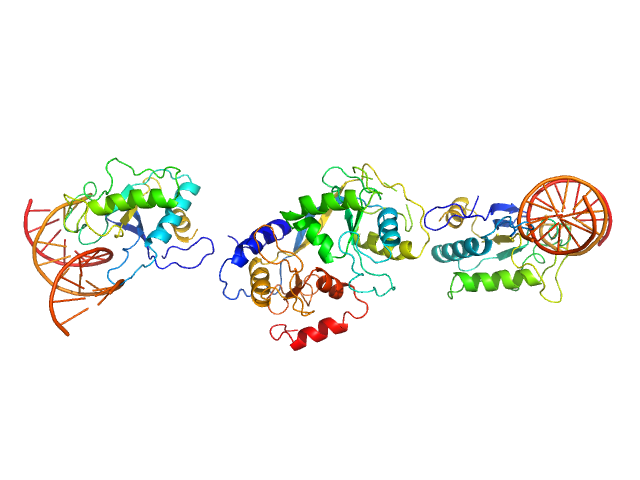
UniProt ID: None (None-None) cognate hemimethylated 12-bp oligoduplex
UniProt ID: P24200 (1-277) 5-methylcytosine-specific restriction enzyme A
|
|
|
|
| Sample: |
Cognate hemimethylated 12-bp oligoduplex dimer, 15 kDa DNA
5-methylcytosine-specific restriction enzyme A dimer, 65 kDa Escherichia coli protein
|
| Buffer: |
20 mM Tris–HCl pH 7.5, 200 mM KCl, 0.1 mM EDTA, 0.01% (w/v) sodium azide and 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 May 24
|
Activity and structure of EcoKMcrA.
Nucleic Acids Res 46(18):9829-9841 (2018)
Czapinska H, Kowalska M, Zagorskaite E, Manakova E, Slyvka A, Xu SY, Siksnys V, Sasnauskas G, Bochtler M
|
| RgGuinier |
3.9 |
nm |
| Dmax |
13.5 |
nm |
| VolumePorod |
69 |
nm3 |
|
|
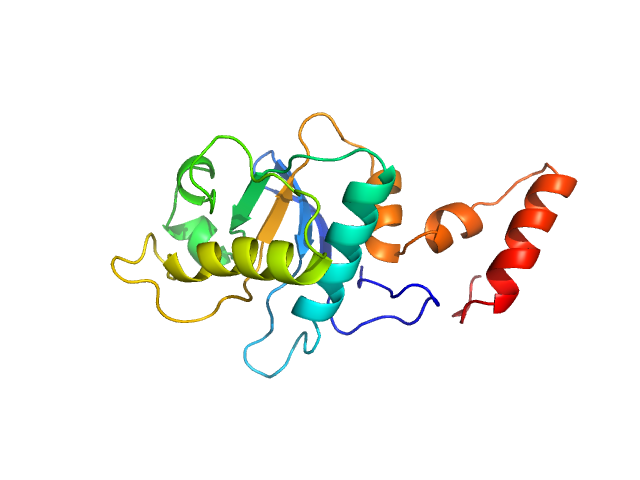
UniProt ID: P24200 (1-175) 5-methylcytosine-specific restriction enzyme A (N-terminal domain)
|
|
|
|
| Sample: |
5-methylcytosine-specific restriction enzyme A (N-terminal domain) monomer, 21 kDa Escherichia coli protein
|
| Buffer: |
20 mM Tris–HCl pH 7.5, 200 mM KCl, 0.1 mM EDTA, 0.01% (w/v) sodium azide, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 May 24
|
Activity and structure of EcoKMcrA.
Nucleic Acids Res 46(18):9829-9841 (2018)
Czapinska H, Kowalska M, Zagorskaite E, Manakova E, Slyvka A, Xu SY, Siksnys V, Sasnauskas G, Bochtler M
|
| RgGuinier |
2.1 |
nm |
| Dmax |
7.5 |
nm |
| VolumePorod |
29 |
nm3 |
|
|
UniProt ID: P24200 (1-175) 5-methylcytosine-specific restriction enzyme A (N-terminal domain)
UniProt ID: None (None-None) cognate hemimethylated 12-bp oligoduplex
|
|
|
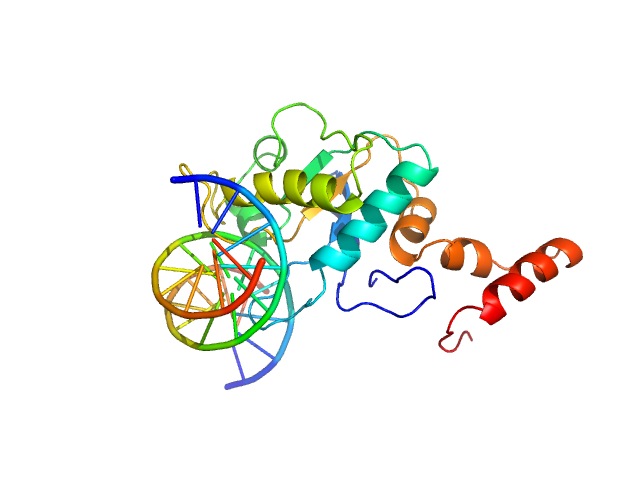
|
| Sample: |
5-methylcytosine-specific restriction enzyme A (N-terminal domain) monomer, 21 kDa Escherichia coli protein
Cognate hemimethylated 12-bp oligoduplex monomer, 7 kDa DNA
|
| Buffer: |
20 mM Tris–HCl pH 7.5, 200 mM KCl, 0.1 mM EDTA, 0.01% (w/v) sodium azide and 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 May 24
|
Activity and structure of EcoKMcrA.
Nucleic Acids Res 46(18):9829-9841 (2018)
Czapinska H, Kowalska M, Zagorskaite E, Manakova E, Slyvka A, Xu SY, Siksnys V, Sasnauskas G, Bochtler M
|
| RgGuinier |
2.1 |
nm |
| Dmax |
7.5 |
nm |
| VolumePorod |
29 |
nm3 |
|
|
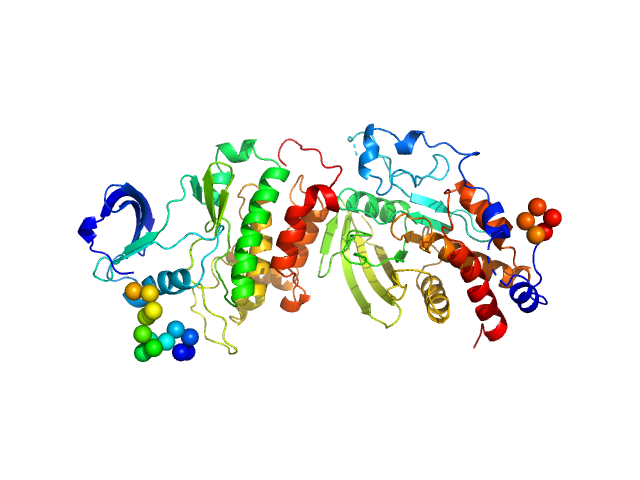
UniProt ID: P23469 (107-400) Receptor-type tyrosine-protein phosphatase epsilon
UniProt ID: P12931 (263-536) Proto-oncogene tyrosine-protein kinase Src, T357M mutant
|
|
|
|
| Sample: |
Receptor-type tyrosine-protein phosphatase epsilon monomer, 34 kDa Homo sapiens protein
Proto-oncogene tyrosine-protein kinase Src, T357M mutant monomer, 31 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris , 50 mM NaCl, 5 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at 23A1, Taiwan Photon Source, NSRRC on 2017 May 26
|
An integrative approach unveils a distal encounter site for rPTPε and phospho-Src complex formation
Structure (2023)
EswarKumar N, Yang C, Tewary S, Peng W, Chen G, Yeh Y, Yang H, Ho M
|
| RgGuinier |
2.9 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
60 |
nm3 |
|
|
UniProt ID: Q8CZ28-1 (1-400) FAD-binding FR-type domain-containing protein
|
|
|
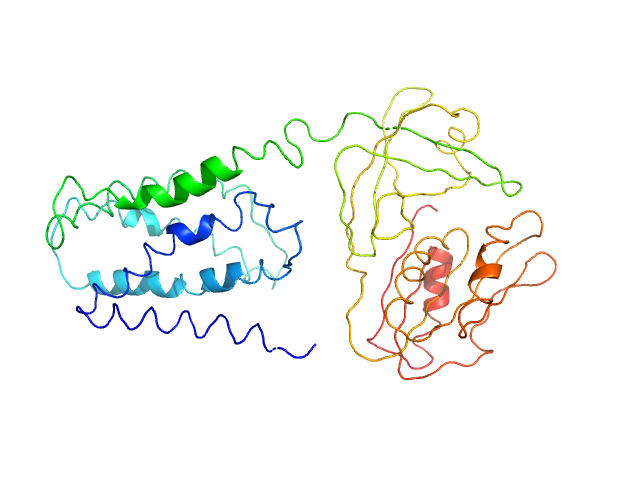
|
| Sample: |
FAD-binding FR-type domain-containing protein monomer, 47 kDa Streptococcus pneumoniae protein
|
| Buffer: |
50 mM Tris-HCl pH 7, 300 mM NaCl, 5 mM LMNG, 10 µM FAD, 21.4% D₂O, pH: 7 |
| Experiment: |
SANS
data collected at D22, Institut Laue-Langevin (ILL) on 2018 Jun 26
|
Interdomain Flexibility within NADPH Oxidase Suggested by SANS Using LMNG Stealth Carrier.
Biophys J 119(3):605-618 (2020)
Vermot A, Petit-Härtlein I, Breyton C, Le Roy A, Thépaut M, Vivès C, Moulin M, Härtlein M, Grudinin S, Smith SME, Ebel C, Martel A, Fieschi F
|
| RgGuinier |
3.0 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
89 |
nm3 |
|
|
UniProt ID: Q96T88 (123-301) E3 ubiquitin-protein ligase UHRF1
|
|
|
|
| Sample: |
E3 ubiquitin-protein ligase UHRF1 monomer, 21 kDa Homo sapiens protein
|
| Buffer: |
SAXS Buffer, pH: 7.5 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2018 Nov 26
|
Serine 298 Phosphorylation in Linker 2 of UHRF1 Regulates Ligand-Binding Property of its Tandem Tudor Domain
Journal of Molecular Biology (2020)
Kori S, Jimenji T, Ekimoto T, Sato M, Kusano F, Oda T, Unoki M, Ikeguchi M, Arita K
|
| RgGuinier |
2.0 |
nm |
| Dmax |
6.6 |
nm |
| VolumePorod |
26 |
nm3 |
|
|
UniProt ID: Q96T88 (123-301) E3 ubiquitin-protein ligase UHRF1
|
|
|
|
| Sample: |
E3 ubiquitin-protein ligase UHRF1 monomer, 21 kDa Homo sapiens protein
|
| Buffer: |
SAXS buffer, pH: 7.5 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2018 Nov 26
|
Serine 298 Phosphorylation in Linker 2 of UHRF1 Regulates Ligand-Binding Property of its Tandem Tudor Domain
Journal of Molecular Biology (2020)
Kori S, Jimenji T, Ekimoto T, Sato M, Kusano F, Oda T, Unoki M, Ikeguchi M, Arita K
|
| RgGuinier |
2.1 |
nm |
| Dmax |
7.0 |
nm |
| VolumePorod |
29 |
nm3 |
|
|
UniProt ID: Q8NBR6 (241-504) Ubiquitin carboxyl-terminal hydrolase MINDY-2
|
|
|
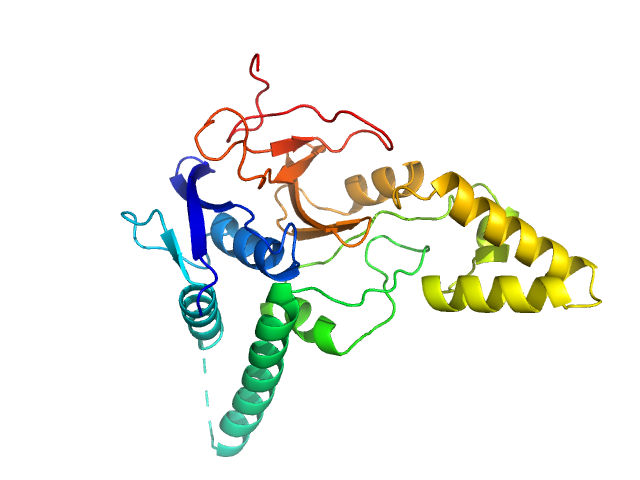
|
| Sample: |
Ubiquitin carboxyl-terminal hydrolase MINDY-2 monomer, 31 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 100 mM NaCl, 5 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Nov 23
|
Mechanism of activation and regulation of deubiquitinase activity in MINDY1 and MINDY2.
Mol Cell (2021)
Abdul Rehman SA, Armstrong LA, Lange SM, Kristariyanto YA, Gräwert TW, Knebel A, Svergun DI, Kulathu Y
|
| RgGuinier |
2.1 |
nm |
| Dmax |
6.5 |
nm |
| VolumePorod |
45 |
nm3 |
|
|
UniProt ID: Q8NBR6 (241-504) Ubiquitin carboxyl-terminal hydrolase MINDY-2
UniProt ID: P0CG48 (1-76) Polyubiquitin-C
|
|
|
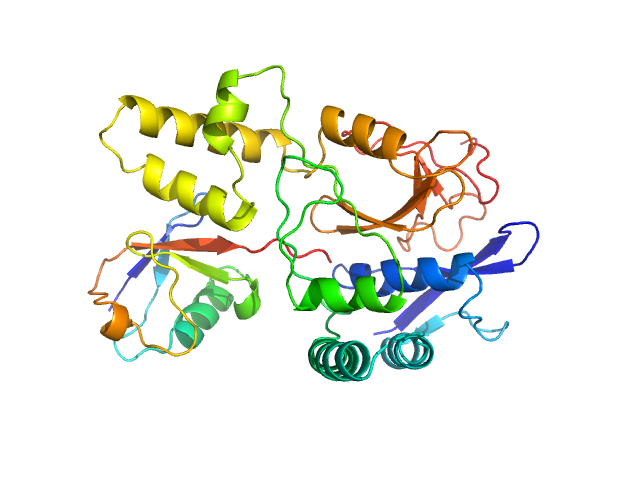
|
| Sample: |
Ubiquitin carboxyl-terminal hydrolase MINDY-2 monomer, 31 kDa Homo sapiens protein
Polyubiquitin-C monomer, 9 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 100 mM NaCl, 5 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Nov 23
|
Mechanism of activation and regulation of deubiquitinase activity in MINDY1 and MINDY2.
Mol Cell (2021)
Abdul Rehman SA, Armstrong LA, Lange SM, Kristariyanto YA, Gräwert TW, Knebel A, Svergun DI, Kulathu Y
|
| RgGuinier |
2.3 |
nm |
| Dmax |
6.8 |
nm |
| VolumePorod |
58 |
nm3 |
|
|
UniProt ID: Q8NBR6 (241-504) Ubiquitin carboxyl-terminal hydrolase C266A mutant
UniProt ID: P0CG48 (1-76) Polyubiquitin-C
|
|
|
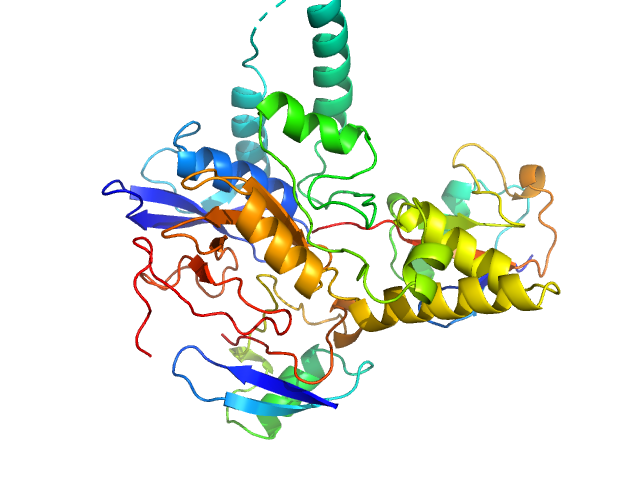
|
| Sample: |
Ubiquitin carboxyl-terminal hydrolase C266A mutant monomer, 31 kDa Homo sapiens protein
Polyubiquitin-C dimer, 17 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 100 mM NaCl, 5 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Nov 23
|
Mechanism of activation and regulation of deubiquitinase activity in MINDY1 and MINDY2.
Mol Cell (2021)
Abdul Rehman SA, Armstrong LA, Lange SM, Kristariyanto YA, Gräwert TW, Knebel A, Svergun DI, Kulathu Y
|
| RgGuinier |
2.5 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
68 |
nm3 |
|
|