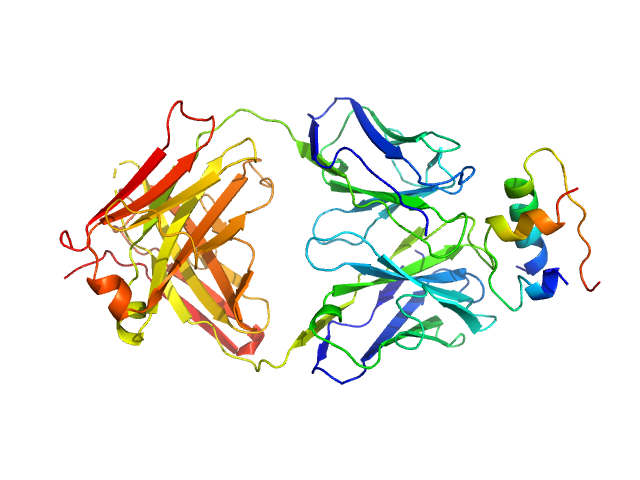
UniProt ID: P01308 (25-110) Insulin (Insulin B chain and Insulin A chain)
UniProt ID: None (None-None) HUI-018 Fab
|
|
|
|
| Sample: |
Insulin (Insulin B chain and Insulin A chain) monomer, 6 kDa Homo sapiens protein
HUI-018 Fab monomer, 47 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES, 140 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at I911-4, MAX IV on 2013 Mar 7
|
Insulin Binding to the Analytical Antibody Sandwich Pair OXI
‐005 and HUI
‐018 – Epitope Mapping and Binding Properties
Protein Science (2020)
Johansson E, Wu X, Yu B, Yang Z, Cao Z, Wiberg C, Jeppesen C, Poulsen F
|
| RgGuinier |
3.4 |
nm |
| Dmax |
10.7 |
nm |
| VolumePorod |
86 |
nm3 |
|
|
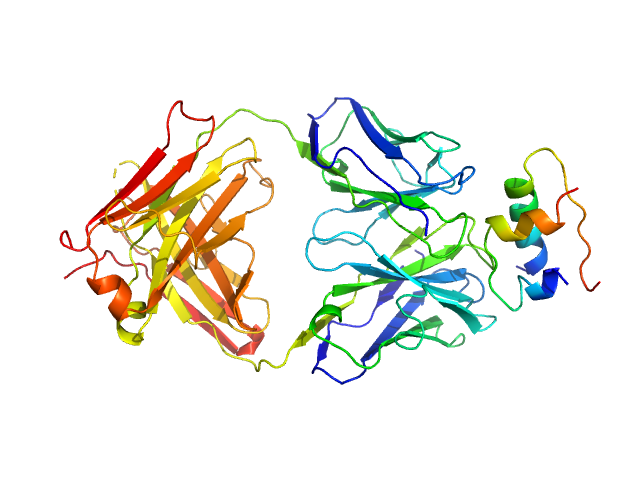
UniProt ID: P01308 (25-110) Insulin (Insulin B chain and Insulin A chain)
UniProt ID: None (None-None) HUI-018 Fab
|
|
|
|
| Sample: |
Insulin (Insulin B chain and Insulin A chain) monomer, 6 kDa Homo sapiens protein
HUI-018 Fab monomer, 47 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES, 140 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at I911-4, MAX IV on 2013 Mar 7
|
Insulin Binding to the Analytical Antibody Sandwich Pair OXI
‐005 and HUI
‐018 – Epitope Mapping and Binding Properties
Protein Science (2020)
Johansson E, Wu X, Yu B, Yang Z, Cao Z, Wiberg C, Jeppesen C, Poulsen F
|
| RgGuinier |
3.7 |
nm |
| Dmax |
13.4 |
nm |
| VolumePorod |
105 |
nm3 |
|
|
UniProt ID: None (None-None) HUI-018 Fab
UniProt ID: P01315 (25-108) Insulin (Insulin B chain and Insulin A chain)
UniProt ID: None (None-None) OXI-005 Fab
|
|
|
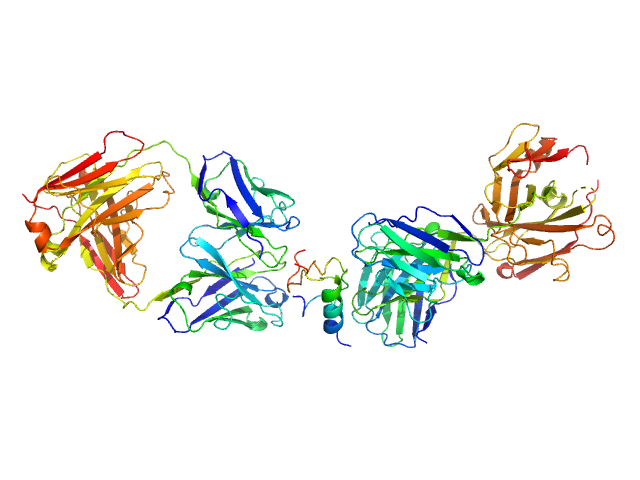
|
| Sample: |
HUI-018 Fab monomer, 47 kDa Mus musculus protein
Insulin (Insulin B chain and Insulin A chain) monomer, 6 kDa Sus scrofa protein
OXI-005 Fab monomer, 47 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES, 140 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-2000, Novo Nordisk A/S on 2019 Mar 28
|
Insulin Binding to the Analytical Antibody Sandwich Pair OXI
‐005 and HUI
‐018 – Epitope Mapping and Binding Properties
Protein Science (2020)
Johansson E, Wu X, Yu B, Yang Z, Cao Z, Wiberg C, Jeppesen C, Poulsen F
|
| RgGuinier |
4.5 |
nm |
| Dmax |
14.2 |
nm |
| VolumePorod |
138 |
nm3 |
|
|
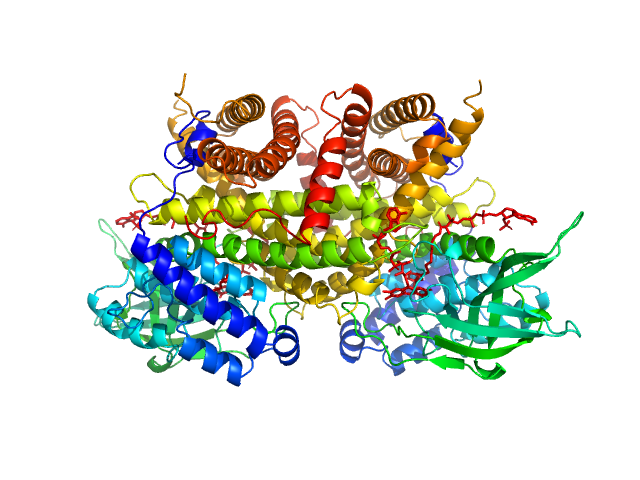
UniProt ID: Q9H845 (38-621) Complex I assembly factor ACAD9, mitochondrial
|
|
|
|
| Sample: |
Complex I assembly factor ACAD9, mitochondrial dimer, 130 kDa Homo sapiens protein
|
| Buffer: |
25mM Tris-HCl, 150mM NaCl, 0.1mM DTT, and 5% glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2017 Nov 2
|
Molecular mechanism of interactions between ACAD9 and binding partners in mitochondrial respiratory complex I assembly
iScience 24(10):103153 (2021)
Xia C, Lou B, Fu Z, Mohsen A, Shen A, Vockley J, Kim J
|
| RgGuinier |
3.5 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
213 |
nm3 |
|
|
UniProt ID: Q9H845 (38-621) Complex I assembly factor ACAD9, mitochondrial
UniProt ID: Q9BQ95 (49-431) Evolutionarily conserved signaling intermediate in Toll pathway, mitochondrial
|
|
|
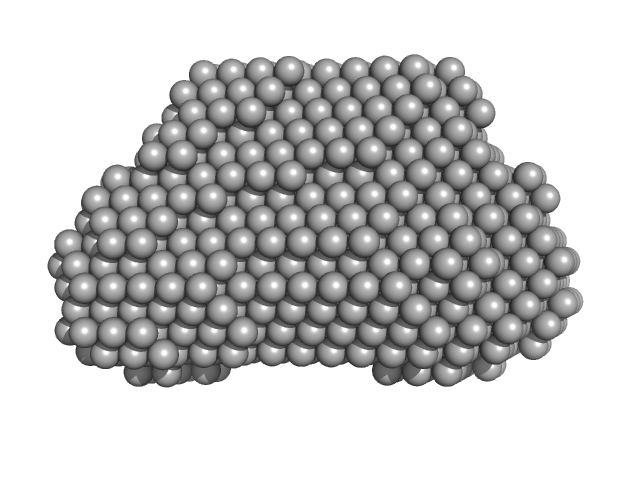
|
| Sample: |
Complex I assembly factor ACAD9, mitochondrial dimer, 130 kDa Homo sapiens protein
Evolutionarily conserved signaling intermediate in Toll pathway, mitochondrial dimer, 93 kDa Homo sapiens protein
|
| Buffer: |
25mM Tris-HCl, 150mM NaCl, 0.1mM DTT, and 5% glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2017 Nov 2
|
Molecular mechanism of interactions between ACAD9 and binding partners in mitochondrial respiratory complex I assembly
iScience 24(10):103153 (2021)
Xia C, Lou B, Fu Z, Mohsen A, Shen A, Vockley J, Kim J
|
| RgGuinier |
5.1 |
nm |
| Dmax |
16.8 |
nm |
| VolumePorod |
514 |
nm3 |
|
|
UniProt ID: Q9H845 (38-621) Complex I assembly factor ACAD9, mitochondrial
UniProt ID: Q9BQ95 (49-431) Evolutionarily conserved signaling intermediate in Toll pathway, mitochondrial
UniProt ID: Q9Y375 (25-327) Complex I intermediate-associated protein 30, mitochondrial
|
|
|

|
| Sample: |
Complex I assembly factor ACAD9, mitochondrial tetramer, 259 kDa Homo sapiens protein
Evolutionarily conserved signaling intermediate in Toll pathway, mitochondrial tetramer, 184 kDa Homo sapiens protein
Complex I intermediate-associated protein 30, mitochondrial tetramer, 190 kDa Homo sapiens protein
|
| Buffer: |
25mM Tris-HCl, 150mM NaCl, 0.1mM DTT, and 5% glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2017 Nov 2
|
Molecular mechanism of interactions between ACAD9 and binding partners in mitochondrial respiratory complex I assembly
iScience 24(10):103153 (2021)
Xia C, Lou B, Fu Z, Mohsen A, Shen A, Vockley J, Kim J
|
| RgGuinier |
6.6 |
nm |
| Dmax |
21.6 |
nm |
| VolumePorod |
1340 |
nm3 |
|
|
UniProt ID: P0DP23 (1-149) Calmodulin-1
|
|
|
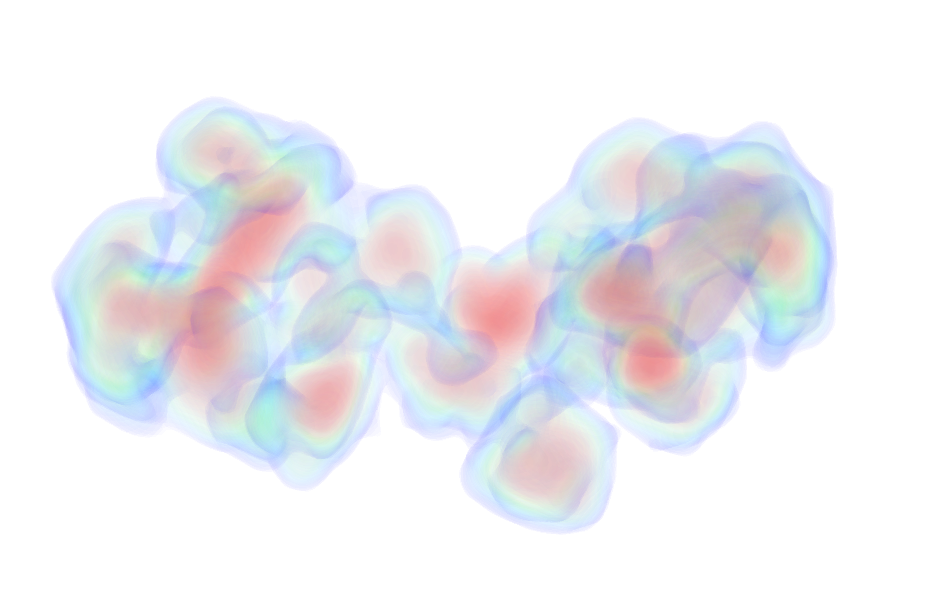
|
| Sample: |
Calmodulin-1 monomer, 17 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 4 mM CaCl₂, pH: 7.4 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2016 Sep 24
|
A High‐Affinity Calmodulin‐Binding Site in the CyaA Toxin Translocation Domain is Essential for Invasion of Eukaryotic Cells
Advanced Science :2003630 (2021)
Voegele A, Sadi M, O'Brien D, Gehan P, Raoux‐Barbot D, Davi M, Hoos S, Brûlé S, Raynal B, Weber P, Mechaly A, Haouz A, Rodriguez N, Vachette P, Durand D, Brier S, Ladant D, Chenal A
|
| RgGuinier |
2.2 |
nm |
| Dmax |
7.3 |
nm |
| VolumePorod |
27 |
nm3 |
|
|
UniProt ID: P0DP23 (1-149) Calmodulin-1
UniProt ID: P0DKX7 (454-484) Bifunctional hemolysin/adenylate cyclase
|
|
|
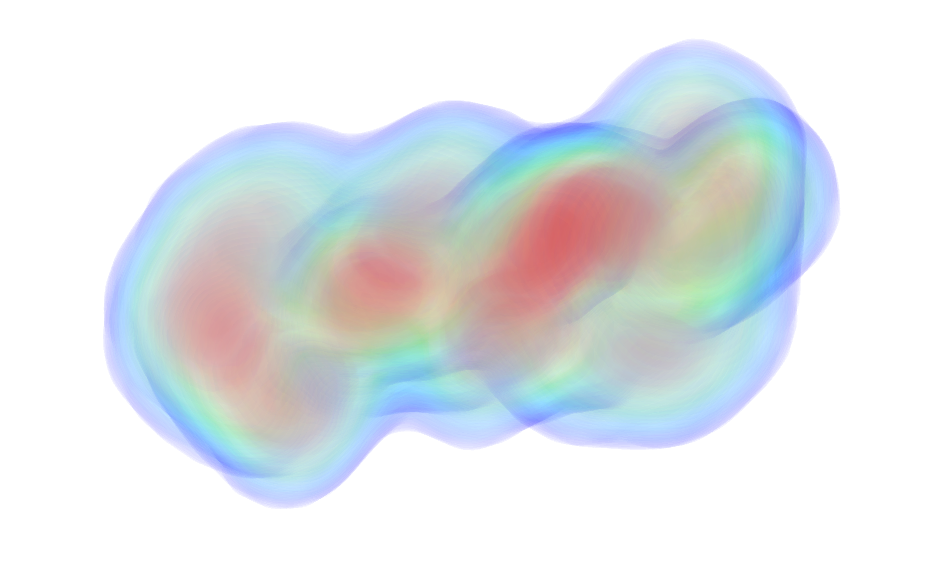
|
| Sample: |
Calmodulin-1 monomer, 17 kDa Homo sapiens protein
Bifunctional hemolysin/adenylate cyclase monomer, 3 kDa Bordetella pertussis protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 4 mM CaCl₂, pH: 7.4 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2016 Sep 24
|
A High‐Affinity Calmodulin‐Binding Site in the CyaA Toxin Translocation Domain is Essential for Invasion of Eukaryotic Cells
Advanced Science :2003630 (2021)
Voegele A, Sadi M, O'Brien D, Gehan P, Raoux‐Barbot D, Davi M, Hoos S, Brûlé S, Raynal B, Weber P, Mechaly A, Haouz A, Rodriguez N, Vachette P, Durand D, Brier S, Ladant D, Chenal A
|
| RgGuinier |
2.0 |
nm |
| Dmax |
7.0 |
nm |
| VolumePorod |
28 |
nm3 |
|
|
UniProt ID: P19809 (450-650) Intimin, D00-D0 domain (6xHis tagged)
|
|
|
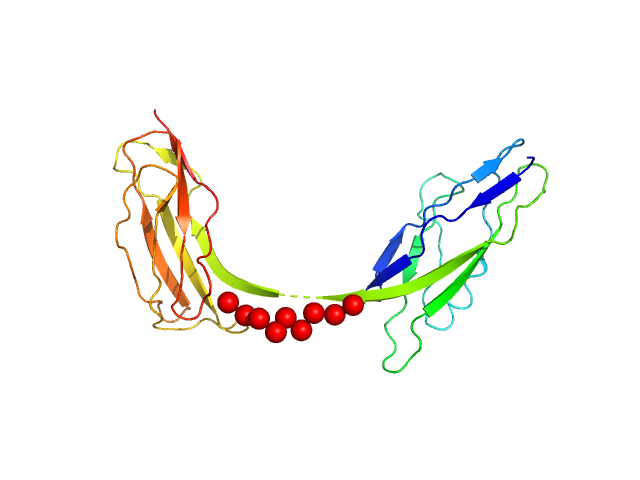
|
| Sample: |
Intimin, D00-D0 domain (6xHis tagged) monomer, 23 kDa Escherichia coli O127:H6 … protein
|
| Buffer: |
10 mM HEPES, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Jul 12
|
The extracellular juncture domains in the intimin passenger adopt a constitutively extended conformation inducing restraints to its sphere of action.
Sci Rep 10(1):21249 (2020)
Weikum J, Kulakova A, Tesei G, Yoshimoto S, Jægerum LV, Schütz M, Hori K, Skepö M, Harris P, Leo JC, Morth JP
|
| RgGuinier |
3.1 |
nm |
| Dmax |
8.3 |
nm |
| VolumePorod |
32 |
nm3 |
|
|
UniProt ID: P19809 (553-752) Intimin (D0-D1 domain, 6xHis tagged)
|
|
|
|
| Sample: |
Intimin (D0-D1 domain, 6xHis tagged) monomer, 23 kDa Escherichia coli O127:H6 … protein
|
| Buffer: |
10 mM HEPES, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Jul 12
|
The extracellular juncture domains in the intimin passenger adopt a constitutively extended conformation inducing restraints to its sphere of action.
Sci Rep 10(1):21249 (2020)
Weikum J, Kulakova A, Tesei G, Yoshimoto S, Jægerum LV, Schütz M, Hori K, Skepö M, Harris P, Leo JC, Morth JP
|
| RgGuinier |
2.2 |
nm |
| Dmax |
5.8 |
nm |
| VolumePorod |
34 |
nm3 |
|
|