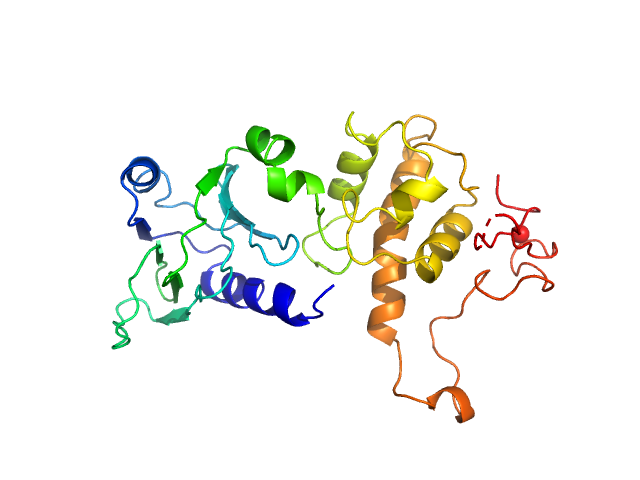
UniProt ID: P50465 (2-263) Endonuclease 8
|
|
|
|
| Sample: |
Endonuclease 8 monomer, 31 kDa Escherichia coli protein
|
| Buffer: |
25 mM Bis-Tris, 150 mM NaCl, 2% glycerol, 1 mM TCEP, pH: 8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 Mar 9
|
Unique Structural Features of Mammalian NEIL2 DNA Glycosylase Prime Its Activity for Diverse DNA Substrates and Environments.
Structure (2020)
Eckenroth BE, Cao VB, Averill AM, Dragon JA, Doublié S
|
| RgGuinier |
2.3 |
nm |
| Dmax |
7.8 |
nm |
| VolumePorod |
45 |
nm3 |
|
|
UniProt ID: F7AMK3 (3-336) Nei like DNA glycosylase 2
|
|
|
|
| Sample: |
Nei like DNA glycosylase 2 monomer, 39 kDa Monodelphis domestica protein
|
| Buffer: |
25 mM Bis-Tris, 150 mM NaCl, 2% glycerol, 1 mM TCEP, pH: 8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 Mar 9
|
Unique Structural Features of Mammalian NEIL2 DNA Glycosylase Prime Its Activity for Diverse DNA Substrates and Environments.
Structure (2020)
Eckenroth BE, Cao VB, Averill AM, Dragon JA, Doublié S
|
| RgGuinier |
2.7 |
nm |
| Dmax |
8.2 |
nm |
| VolumePorod |
58 |
nm3 |
|
|
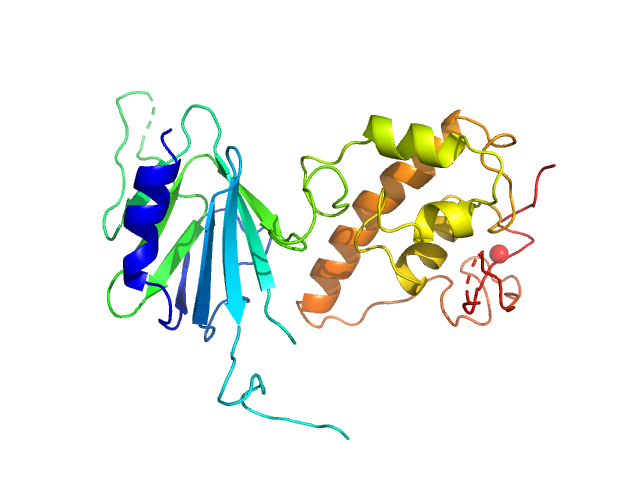
UniProt ID: F7AMK3 (None-None) Nei like DNA glycosylase 2 (Δ67-133)
|
|
|
|
| Sample: |
Nei like DNA glycosylase 2 (Δ67-133) monomer, 33 kDa Monodelphis domestica protein
|
| Buffer: |
25 mM Bis-Tris, 150 mM NaCl, 2% glycerol, 1 mM TCEP, pH: 8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 Mar 9
|
Unique Structural Features of Mammalian NEIL2 DNA Glycosylase Prime Its Activity for Diverse DNA Substrates and Environments.
Structure (2020)
Eckenroth BE, Cao VB, Averill AM, Dragon JA, Doublié S
|
| RgGuinier |
2.4 |
nm |
| Dmax |
7.8 |
nm |
| VolumePorod |
47 |
nm3 |
|
|
UniProt ID: P02945 (14-262) Bacteriorhodopsin
|
|
|
|
| Sample: |
Bacteriorhodopsin monomer, 27 kDa Halobacterium salinarum protein
|
| Buffer: |
25 mM NaH2PO4, 1.35 mM KOH, 40 mM partially-deuterated octyl glucoside mixture, pH: 5.6 |
| Experiment: |
SANS
data collected at NGB 30m SANS, NIST Center for High Resolution Neutron Scattering (CHRNS) on 2017 Jan 20
|
Direct localization of detergents and bacteriorhodopsin in the lipidic cubic phase by small-angle neutron scattering
IUCrJ 8(1) (2021)
Cleveland IV T, Blick E, Krueger S, Leung A, Darwish T, Butler P
|
| RgGuinier |
2.7 |
nm |
| Dmax |
5.4 |
nm |
| VolumePorod |
68 |
nm3 |
|
|
UniProt ID: P02945 (14-262) Bacteriorhodopsin
|
|
|
|
| Sample: |
Bacteriorhodopsin monomer, 27 kDa Halobacterium salinarum protein
|
| Buffer: |
25 mM NaH2PO4, 1.35 mM KOH, 40 mM octyl glucoside, pH: 5.6 |
| Experiment: |
SANS
data collected at NG7, NIST Center for High Resolution Neutron Scattering (CHRNS) on 2017 May 15
|
Direct localization of detergents and bacteriorhodopsin in the lipidic cubic phase by small-angle neutron scattering
IUCrJ 8(1) (2021)
Cleveland IV T, Blick E, Krueger S, Leung A, Darwish T, Butler P
|
| RgGuinier |
2.7 |
nm |
| Dmax |
6.2 |
nm |
| VolumePorod |
49 |
nm3 |
|
|
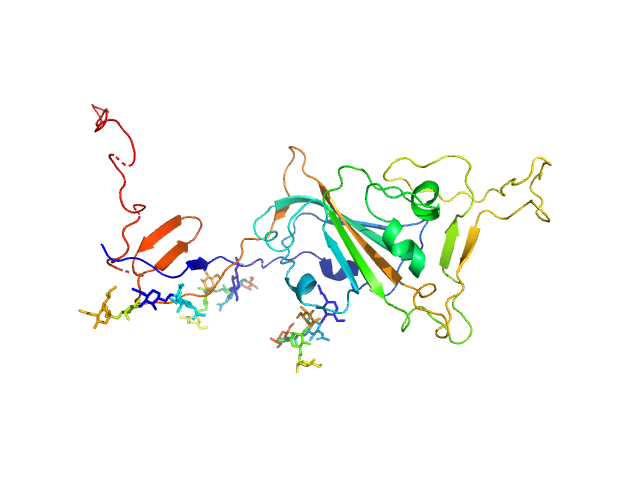
UniProt ID: P0DTC2 (319-566) Spike glycoprotein (ACE2 receptor binding domain)
|
|
|
|
| Sample: |
Spike glycoprotein (ACE2 receptor binding domain) monomer, 29 kDa Severe acute respiratory … protein
|
| Buffer: |
25 mM Tris 100 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 May 1
|
Selection, biophysical and structural analysis of synthetic nanobodies that effectively neutralize SARS-CoV-2
Nature Communications 11(1) (2020)
Custódio T, Das H, Sheward D, Hanke L, Pazicky S, Pieprzyk J, Sorgenfrei M, Schroer M, Gruzinov A, Jeffries C, Graewert M, Svergun D, Dobrev N, Remans K, Seeger M, McInerney G, Murrell B, Hällberg B, Löw C
|
| RgGuinier |
3.0 |
nm |
| Dmax |
13.1 |
nm |
| VolumePorod |
64 |
nm3 |
|
|
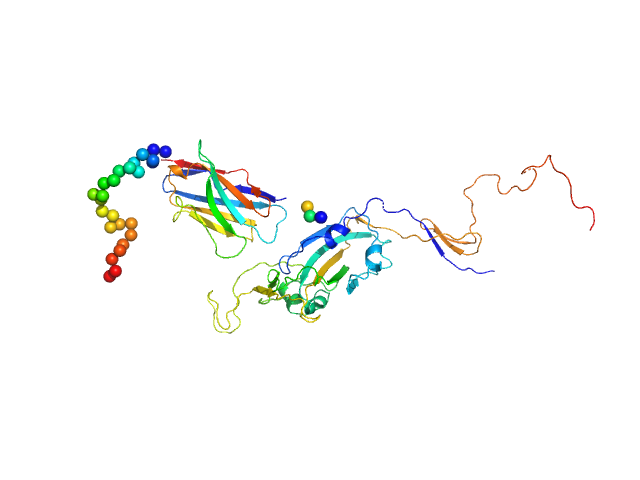
UniProt ID: None (None-None) Synthetic nanobody Sybody 23
UniProt ID: P0DTC2 (319-566) Spike glycoprotein (ACE2 receptor binding domain)
|
|
|
|
| Sample: |
Synthetic nanobody Sybody 23 monomer, 16 kDa synthetic construct protein
Spike glycoprotein (ACE2 receptor binding domain) monomer, 29 kDa Severe acute respiratory … protein
|
| Buffer: |
25 mM Tris 100 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 May 10
|
Selection, biophysical and structural analysis of synthetic nanobodies that effectively neutralize SARS-CoV-2
Nature Communications 11(1) (2020)
Custódio T, Das H, Sheward D, Hanke L, Pazicky S, Pieprzyk J, Sorgenfrei M, Schroer M, Gruzinov A, Jeffries C, Graewert M, Svergun D, Dobrev N, Remans K, Seeger M, McInerney G, Murrell B, Hällberg B, Löw C
|
| RgGuinier |
3.5 |
nm |
| Dmax |
15.1 |
nm |
| VolumePorod |
87 |
nm3 |
|
|
UniProt ID: P9WGZ1 (1-347) DNA-directed RNA polymerase subunit alpha
UniProt ID: P9WGY9 (7-1178) DNA-directed RNA polymerase subunit beta
UniProt ID: P9WGY7 (2-1316) DNA-directed RNA polymerase subunit beta'
UniProt ID: P9WGY5 (2-110) DNA-directed RNA polymerase subunit omega
|
|
|

|
| Sample: |
DNA-directed RNA polymerase subunit alpha dimer, 78 kDa Mycobacterium tuberculosis protein
DNA-directed RNA polymerase subunit beta monomer, 129 kDa Mycobacterium tuberculosis protein
DNA-directed RNA polymerase subunit beta' monomer, 147 kDa Mycobacterium tuberculosis protein
DNA-directed RNA polymerase subunit omega monomer, 12 kDa Mycobacterium tuberculosis protein
|
| Buffer: |
10 mM Tris-Cl, 100 mM NaCl, 10 mM MgCl2, 0.1 mM EDTA, 5% Glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Mar 8
|
Structural and biochemical characterization of RNA polymerase core and UvrD complex: a key component in transcription coupled DNA repair
Ravishankar Ramachandran
|
| RgGuinier |
5.1 |
nm |
| Dmax |
19.3 |
nm |
| VolumePorod |
262 |
nm3 |
|
|
UniProt ID: P9WGZ1 (1-347) DNA-directed RNA polymerase subunit alpha
UniProt ID: P9WGY9 (7-1178) DNA-directed RNA polymerase subunit beta
UniProt ID: P9WGY7 (2-1316) DNA-directed RNA polymerase subunit beta'
UniProt ID: P9WGY5 (1-110) DNA-directed RNA polymerase subunit omega
UniProt ID: P9WMQ1 (1-771) ATP-dependent DNA helicase UvrD1
|
|
|

|
| Sample: |
DNA-directed RNA polymerase subunit alpha dimer, 75 kDa Mycobacterium tuberculosis protein
DNA-directed RNA polymerase subunit beta monomer, 129 kDa Mycobacterium tuberculosis protein
DNA-directed RNA polymerase subunit beta' monomer, 147 kDa Mycobacterium tuberculosis protein
DNA-directed RNA polymerase subunit omega monomer, 12 kDa Mycobacterium tuberculosis protein
ATP-dependent DNA helicase UvrD1 monomer, 85 kDa Mycobacterium tuberculosis protein
|
| Buffer: |
10 mM Tris-Cl, 100 mM NaCl, 10 mM MgCl2, 0.1 mM EDTA, 5% Glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Mar 11
|
Structural and biochemical characterization of RNA polymerase core and UvrD complex: a key component in transcription coupled DNA repair
Ravishankar Ramachandran
|
| RgGuinier |
7.5 |
nm |
| Dmax |
22.2 |
nm |
| VolumePorod |
898 |
nm3 |
|
|
UniProt ID: P9WMQ1 (1-771) ATP-dependent DNA helicase UvrD1
|
|
|
|
| Sample: |
ATP-dependent DNA helicase UvrD1 monomer, 85 kDa Mycobacterium tuberculosis protein
|
| Buffer: |
10 mM Tris-Cl, 100 mM NaCl, 10 mM MgCl2, 0.1 mM EDTA, 5% Glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpace, CSIR-Central Drug Research Institute on 2020 Feb 28
|
Structural and biochemical characterization of RNA polymerase core and UvrD complex: a key component in transcription coupled DNA repair
Ravishankar Ramachandran
|
| RgGuinier |
3.4 |
nm |
| Dmax |
8.4 |
nm |
| VolumePorod |
132 |
nm3 |
|
|