UniProt ID: P01023 (24-1474) Alpha-2-macroglobulin
|
|
|
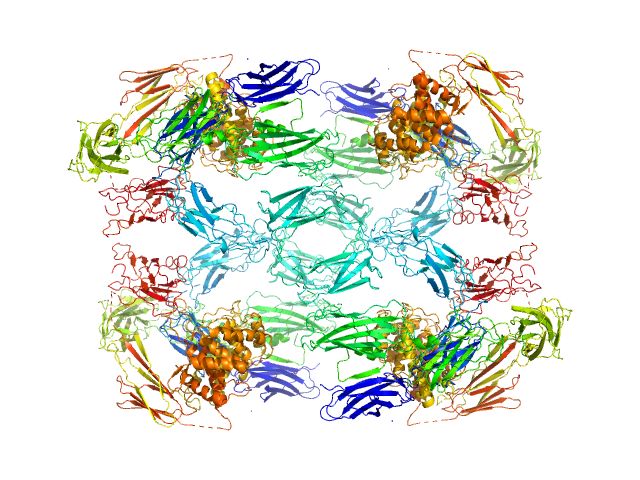
|
| Sample: |
Alpha-2-macroglobulin tetramer, 643 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at Bruker Nanostar w Excillum source, Department of Chemistry, iNANO building, Aarhus Uinversity on 2019 Apr 12
|
Structural Investigations of Human A2M Identify a Hollow Native Conformation That Underlies Its Distinctive Protease-Trapping Mechanism.
Mol Cell Proteomics 20:100090 (2021)
Harwood SL, Lyngsø J, Zarantonello A, Kjøge K, Nielsen PK, Andersen GR, Pedersen JS, Enghild JJ
|
| RgGuinier |
7.6 |
nm |
| Dmax |
20.2 |
nm |
|
|
UniProt ID: P01023 (24-1474) Alpha-2-macroglobulin
|
|
|
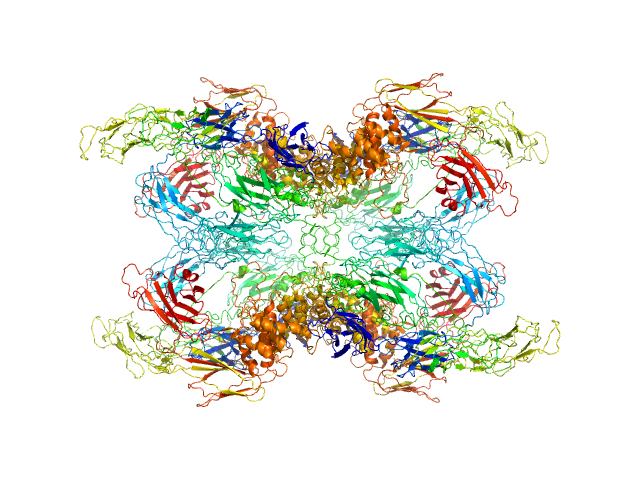
|
| Sample: |
Alpha-2-macroglobulin tetramer, 643 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at Bruker Nanostar w Excillum source, Department of Chemistry, iNANO building, Aarhus Uinversity on 2019 Apr 13
|
Structural Investigations of Human A2M Identify a Hollow Native Conformation That Underlies Its Distinctive Protease-Trapping Mechanism.
Mol Cell Proteomics 20:100090 (2021)
Harwood SL, Lyngsø J, Zarantonello A, Kjøge K, Nielsen PK, Andersen GR, Pedersen JS, Enghild JJ
|
| RgGuinier |
6.6 |
nm |
| Dmax |
19.7 |
nm |
|
|
UniProt ID: P00760 (1-246) Cationic trypsin
UniProt ID: P01023 (24-1474) Alpha-2-macroglobulin
|
|
|
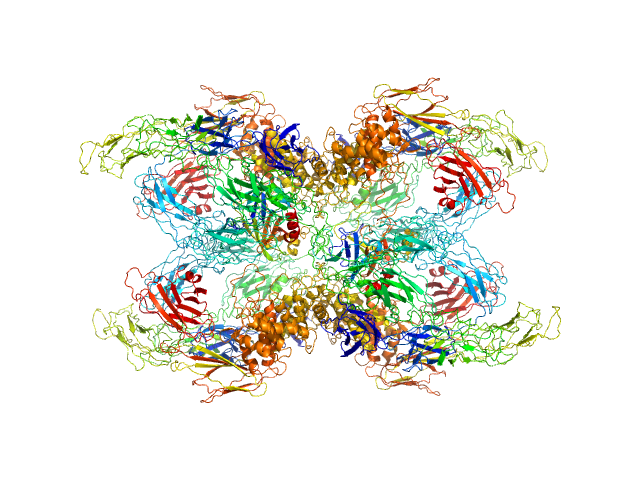
|
| Sample: |
Cationic trypsin dimer, 52 kDa Bos taurus protein
Alpha-2-macroglobulin tetramer, 643 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at Bruker Nanostar w Excillum source, Department of Chemistry, iNANO building, Aarhus Uinversity on 2020 Jan 22
|
Structural Investigations of Human A2M Identify a Hollow Native Conformation That Underlies Its Distinctive Protease-Trapping Mechanism.
Mol Cell Proteomics 20:100090 (2021)
Harwood SL, Lyngsø J, Zarantonello A, Kjøge K, Nielsen PK, Andersen GR, Pedersen JS, Enghild JJ
|
| RgGuinier |
6.6 |
nm |
| Dmax |
20.7 |
nm |
|
|
UniProt ID: Q8DZX1 (1-250) ABC transporter, ATP-binding protein (Nucleotide-Binding Domain SaNsrF)
|
|
|
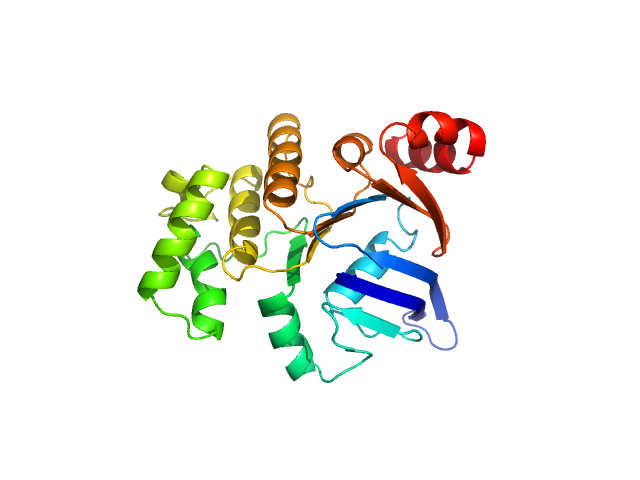
|
| Sample: |
ABC transporter, ATP-binding protein (Nucleotide-Binding Domain SaNsrF) monomer, 31 kDa Streptococcus agalactiae protein
|
| Buffer: |
100 mM HEPES, 150 mM NaCl, 10% glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at Xenocs Xeuss 2.0 Q-Xoom, Center for Structural Studies, Heinrich-Heine-University on 2019 Dec 11
|
Characterization of the nucleotide-binding domain NsrF from the BceAB-type ABC-transporter NsrFP from the human pathogen Streptococcus agalactiae
Scientific Reports 10(1) (2020)
Furtmann F, Porta N, Hoang D, Reiners J, Schumacher J, Gottstein J, Gohlke H, Smits S
|
| RgGuinier |
2.4 |
nm |
| Dmax |
7.9 |
nm |
| VolumePorod |
64 |
nm3 |
|
|
UniProt ID: P0DKX7 (1132-1681) Bifunctional hemolysin/adenylate cyclase
|
|
|

|
| Sample: |
Bifunctional hemolysin/adenylate cyclase monomer, 57 kDa Bordetella pertussis protein
|
| Buffer: |
10 mM Tris HCl, 150 mM NaCl, 10 mM CaCl₂, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Oct 31
|
Continuous Assembly of β-Roll Structures Is Implicated in the Type I-Dependent Secretion of Large Repeat-in-Toxins (RTX) Proteins.
J Mol Biol (2020)
Motlova L, Klimova N, Fiser R, Sebo P, Bumba L
|
| RgGuinier |
4.0 |
nm |
| Dmax |
13.3 |
nm |
| VolumePorod |
94 |
nm3 |
|
|
UniProt ID: P0DKX7 (1243-1681) Bifunctional hemolysin/adenylate cyclase
|
|
|

|
| Sample: |
Bifunctional hemolysin/adenylate cyclase monomer, 45 kDa Bordetella pertussis protein
|
| Buffer: |
10 mM Tris HCl, 150 mM NaCl, 10 mM CaCl₂, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Oct 31
|
Continuous Assembly of β-Roll Structures Is Implicated in the Type I-Dependent Secretion of Large Repeat-in-Toxins (RTX) Proteins.
J Mol Biol (2020)
Motlova L, Klimova N, Fiser R, Sebo P, Bumba L
|
| RgGuinier |
3.4 |
nm |
| Dmax |
12.1 |
nm |
| VolumePorod |
85 |
nm3 |
|
|
UniProt ID: P0DKX7 (1372-1681) Bifunctional hemolysin/adenylate cyclase
|
|
|

|
| Sample: |
Bifunctional hemolysin/adenylate cyclase monomer, 32 kDa Bordetella pertussis protein
|
| Buffer: |
10 mM Tris HCl, 150 mM NaCl, 10 mM CaCl₂, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Oct 31
|
Continuous Assembly of β-Roll Structures Is Implicated in the Type I-Dependent Secretion of Large Repeat-in-Toxins (RTX) Proteins.
J Mol Biol (2020)
Motlova L, Klimova N, Fiser R, Sebo P, Bumba L
|
| RgGuinier |
2.7 |
nm |
| Dmax |
8.6 |
nm |
| VolumePorod |
45 |
nm3 |
|
|
UniProt ID: A0A655XAV0 (73-204) Outer membrane virulence protein yopE
|
|
|
|
| Sample: |
Outer membrane virulence protein yopE monomer, 15 kDa Vibrio cholerae protein
|
| Buffer: |
10 mM HEPES-HCl, pH 7.4, 150 mM NaCl, 0.5 mM TCEP, pH: 7.4 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2016 Jun 29
|
Solution structure and dynamics of the mitochondrial‐targeted GTPase
‐activating protein ( GAP
) VopE
by an integrated NMR
/ SAXS
approach
Protein Science (2022)
Smith K, Lee W, Tonelli M, Lee Y, Light S, Cornilescu G, Chakravarthy S
|
| RgGuinier |
2.0 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
28 |
nm3 |
|
|
UniProt ID: V5VFJ0 (20-235) BON domain protein
|
|
|

|
| Sample: |
BON domain protein decamer, 229 kDa Acinetobacter baumannii protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 0.03 % NaN3, 5.0 % glycerol, pH: 7.8 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2017 Apr 11
|
BonA from Acinetobacter baumannii Forms a Divisome-Localized Decamer That Supports Outer Envelope Function.
mBio :e0148021 (2021)
Grinter R, Morris FC, Dunstan RA, Leung PM, Kropp A, Belousoff M, Gunasinghe SD, Scott NE, Beckham S, Peleg AY, Greening C, Li J, Heinz E, Lithgow T
|
| RgGuinier |
4.7 |
nm |
| Dmax |
16.4 |
nm |
| VolumePorod |
546 |
nm3 |
|
|
UniProt ID: V5VFJ0 (46-235) BON domain protein
|
|
|

|
| Sample: |
BON domain protein monomer, 20 kDa Acinetobacter baumannii protein
|
| Buffer: |
20 mM Tris HCl, 150 nM NaCl, 0.02 % NaN3, 5% glycerol, pH: 7.8 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2017 Apr 11
|
BonA from Acinetobacter baumannii Forms a Divisome-Localized Decamer That Supports Outer Envelope Function.
mBio :e0148021 (2021)
Grinter R, Morris FC, Dunstan RA, Leung PM, Kropp A, Belousoff M, Gunasinghe SD, Scott NE, Beckham S, Peleg AY, Greening C, Li J, Heinz E, Lithgow T
|
| RgGuinier |
3.1 |
nm |
| Dmax |
10.8 |
nm |
| VolumePorod |
49 |
nm3 |
|
|