UniProt ID: P0DKX7 (None-None) Adenylate cyclase toxin Block I-V
|
|
|

|
| Sample: |
Adenylate cyclase toxin Block I-V monomer, 70 kDa Bordetella pertussis protein
|
| Buffer: |
10 mM Tris HCl 150 mM NaCl 10 mM CaCl2, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Oct 31
|
Calcium-Driven Folding of RTX Domain β-Rolls Ratchets Translocation of RTX Proteins through Type I Secretion Ducts.
Mol Cell 62(1):47-62 (2016)
Bumba L, Masin J, Macek P, Wald T, Motlova L, Bibova I, Klimova N, Bednarova L, Veverka V, Kachala M, Svergun DI, Barinka C, Sebo P
|
| RgGuinier |
5.6 |
nm |
| Dmax |
17.2 |
nm |
| VolumePorod |
275 |
nm3 |
|
|
UniProt ID: P0DKX7 (None-None) Adenylate cyclase toxin Block V
|
|
|

|
| Sample: |
Adenylate cyclase toxin Block V monomer, 16 kDa Bordetella pertussis protein
|
| Buffer: |
10 mM Tris HCl 150 mM NaCl 10 mM CaCl2, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Oct 31
|
Calcium-Driven Folding of RTX Domain β-Rolls Ratchets Translocation of RTX Proteins through Type I Secretion Ducts.
Mol Cell 62(1):47-62 (2016)
Bumba L, Masin J, Macek P, Wald T, Motlova L, Bibova I, Klimova N, Bednarova L, Veverka V, Kachala M, Svergun DI, Barinka C, Sebo P
|
| RgGuinier |
1.8 |
nm |
| Dmax |
5.9 |
nm |
| VolumePorod |
24 |
nm3 |
|
|
UniProt ID: A0A1Z1SYD5 (64-243) C-terminal catalytic domain of Suppressor of Copper Sensitivity C protein
|
|
|
|
| Sample: |
C-terminal catalytic domain of Suppressor of Copper Sensitivity C protein monomer, 20 kDa Proteus mirabilis protein
|
| Buffer: |
25 mM HEPES 150mM NaCl 1mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2012 Feb 29
|
A shape-shifting redox foldase contributes to Proteus mirabilis copper resistance.
Nat Commun 8:16065 (2017)
Furlong EJ, Lo AW, Kurth F, Premkumar L, Totsika M, Achard MES, Halili MA, Heras B, Whitten AE, Choudhury HG, Schembri MA, Martin JL
|
| RgGuinier |
3.7 |
nm |
| Dmax |
10.5 |
nm |
| VolumePorod |
92 |
nm3 |
|
|
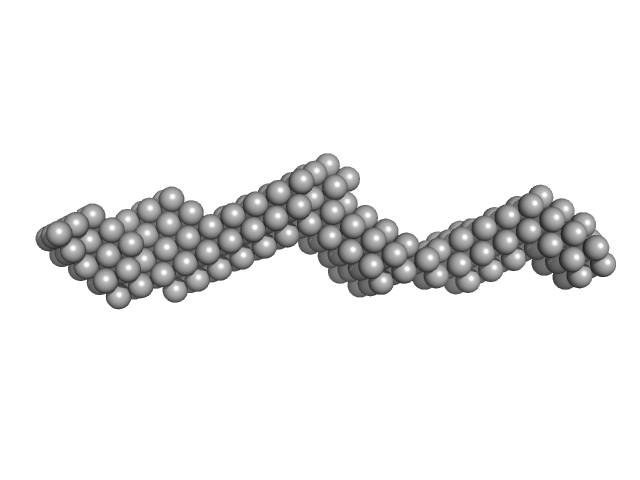
UniProt ID: Q15149-2 (543-1006) Plakin domain fragment of Human plectin encompassing spectrin repeats SR3-SR4-SR5-SR6 and SH3
|
|
|
|
| Sample: |
Plakin domain fragment of Human plectin encompassing spectrin repeats SR3-SR4-SR5-SR6 and SH3 monomer, 53 kDa Homo sapiens protein
|
| Buffer: |
20 mM Sodium Phosphate 150 mM NaCl 5% glycerol 3 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Nov 26
|
The Structure of the Plakin Domain of Plectin Reveals an Extended Rod-like Shape.
J Biol Chem 291(36):18643-62 (2016)
Ortega E, Manso JA, Buey RM, Carballido AM, Carabias A, Sonnenberg A, de Pereda JM
|
| RgGuinier |
5.1 |
nm |
| Dmax |
21.0 |
nm |
| VolumePorod |
91 |
nm3 |
|
|
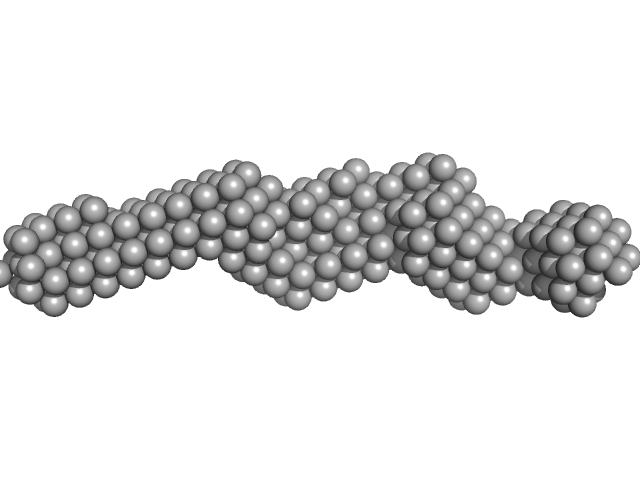
UniProt ID: Q15149-2 (1004-1372) Plakin domain fragment of Human plectin encompassing spectrin repeats SR7-SR8-SR9
|
|
|
|
| Sample: |
Plakin domain fragment of Human plectin encompassing spectrin repeats SR7-SR8-SR9 monomer, 43 kDa Homo sapiens protein
|
| Buffer: |
20 mM Sodium Phosphate 150 mM NaCl 5% glycerol 3 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Nov 26
|
The Structure of the Plakin Domain of Plectin Reveals an Extended Rod-like Shape.
J Biol Chem 291(36):18643-62 (2016)
Ortega E, Manso JA, Buey RM, Carballido AM, Carabias A, Sonnenberg A, de Pereda JM
|
| RgGuinier |
4.2 |
nm |
| Dmax |
17.0 |
nm |
| VolumePorod |
57 |
nm3 |
|
|
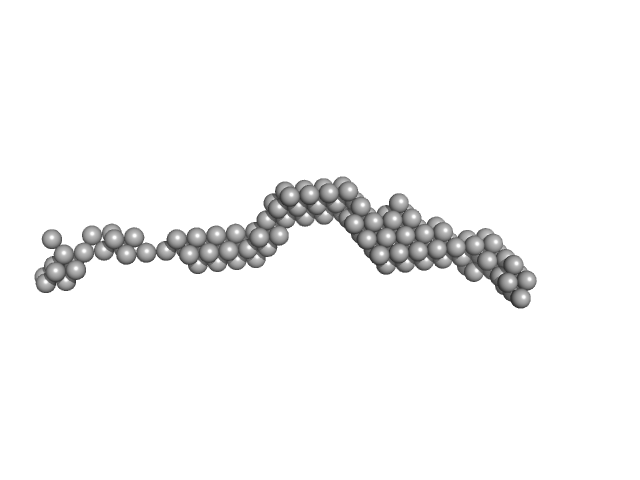
UniProt ID: Q15149-2 (543-1372) Plakin domain fragment of Human plectin encompassing spectrin repeats SR3-SR9
|
|
|
|
| Sample: |
Plakin domain fragment of Human plectin encompassing spectrin repeats SR3-SR9 monomer, 96 kDa Homo sapiens protein
|
| Buffer: |
20 mM Sodium Phosphate 150 mM NaCl 5% glycerol 3 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Aug 13
|
The Structure of the Plakin Domain of Plectin Reveals an Extended Rod-like Shape.
J Biol Chem 291(36):18643-62 (2016)
Ortega E, Manso JA, Buey RM, Carballido AM, Carabias A, Sonnenberg A, de Pereda JM
|
| RgGuinier |
8.5 |
nm |
| Dmax |
35.0 |
nm |
| VolumePorod |
135 |
nm3 |
|
|
UniProt ID: P15924 (660-1025) Plakin domain fragment of Human Desmoplakin encompassing spectrin repeats SR7-SR8-SR9
|
|
|

|
| Sample: |
Plakin domain fragment of Human Desmoplakin encompassing spectrin repeats SR7-SR8-SR9 monomer, 42 kDa Homo sapiens protein
|
| Buffer: |
20 mM Sodium Phosphate 150 mM NaCl 5% glycerol 3 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2015 Sep 25
|
The Structure of the Plakin Domain of Plectin Reveals an Extended Rod-like Shape.
J Biol Chem 291(36):18643-62 (2016)
Ortega E, Manso JA, Buey RM, Carballido AM, Carabias A, Sonnenberg A, de Pereda JM
|
| RgGuinier |
4.4 |
nm |
| Dmax |
17.5 |
nm |
| VolumePorod |
69 |
nm3 |
|
|
UniProt ID: P15924 (180-1025) Plakin domain of Human Desmoplakin
|
|
|
|
| Sample: |
Plakin domain of Human Desmoplakin monomer, 99 kDa Homo sapiens protein
|
| Buffer: |
20 mM Sodium Phosphate 150 mM NaCl 5% glycerol 3 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2015 Sep 25
|
The Structure of the Plakin Domain of Plectin Reveals an Extended Rod-like Shape.
J Biol Chem 291(36):18643-62 (2016)
Ortega E, Manso JA, Buey RM, Carballido AM, Carabias A, Sonnenberg A, de Pereda JM
|
| RgGuinier |
7.5 |
nm |
| Dmax |
35.0 |
nm |
| VolumePorod |
136 |
nm3 |
|
|
UniProt ID: (None-None) short-crRNA: CRISPR/Cas Type I-F Cascade component
UniProt ID: A4Y6G3 (1-183) Cas6f: CRISPR/Cas Type I-F Cascade component (CRISPR-associated protein, Csy4 family)
UniProt ID: A4Y6G1 (1-315) Trimeric Cas7fv: CRISPR/Cas Type I-F Cascade component (Uncharacterized protein, Sputcn32_1821)
UniProt ID: A4Y6G2 (1-336) Cas5fv: CRISPR/Cas Type I-F Cascade component (Uncharacterized protein, Sputcn32_1822)
|
|
|

|
| Sample: |
Short-crRNA: CRISPR/Cas Type I-F Cascade component monomer, 14 kDa Shewanella putrefaciens RNA
Cas6f: CRISPR/Cas Type I-F Cascade component (CRISPR-associated protein, Csy4 family) monomer, 21 kDa Shewanella putrefaciens protein
Trimeric Cas7fv: CRISPR/Cas Type I-F Cascade component (Uncharacterized protein, Sputcn32_1821) trimer, 112 kDa Shewanella putrefaciens protein
Cas5fv: CRISPR/Cas Type I-F Cascade component (Uncharacterized protein, Sputcn32_1822) monomer, 38 kDa Shewanella putrefaciens protein
|
| Buffer: |
50 mM HEPES 150 mM NaCl 1mM DTT 1mM EDTA, pH: 7 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2015 Jun 27
|
Modulating the Cascade architecture of a minimal Type I-F CRISPR-Cas system.
Nucleic Acids Res 44(12):5872-82 (2016)
Gleditzsch D, Müller-Esparza H, Pausch P, Sharma K, Dwarakanath S, Urlaub H, Bange G, Randau L
|
| RgGuinier |
4.1 |
nm |
| Dmax |
14.2 |
nm |
|
|
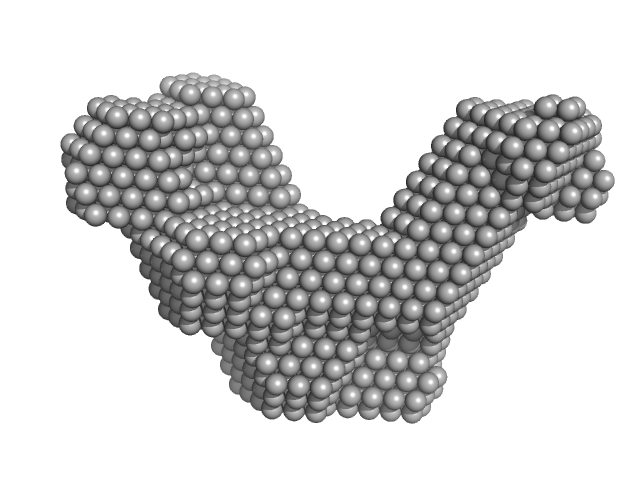
UniProt ID: A4Y6G3 (1-183) Cas6f: CRISPR/Cas Type I-F Cascade component (CRISPR-associated protein, Csy4 family)
UniProt ID: A4Y6G2 (1-336) Cas5fv: CRISPR/Cas Type I-F Cascade component (Uncharacterized protein, Sputcn32_1822)
UniProt ID: A4Y6G1 (1-315) Hexameric Cas7fv: CRISPR/Cas Type I-F Cascade component (Uncharacterized protein, Sputcn32_1821)
UniProt ID: (None-None) wildtype-crRNA: CRISPR/Cas Type I-F Cascade component
|
|
|
|
| Sample: |
Cas6f: CRISPR/Cas Type I-F Cascade component (CRISPR-associated protein, Csy4 family) monomer, 21 kDa Shewanella putrefaciens protein
Cas5fv: CRISPR/Cas Type I-F Cascade component (Uncharacterized protein, Sputcn32_1822) monomer, 38 kDa Shewanella putrefaciens protein
Hexameric Cas7fv: CRISPR/Cas Type I-F Cascade component (Uncharacterized protein, Sputcn32_1821) hexamer, 223 kDa Shewanella putrefaciens protein
Wildtype-crRNA: CRISPR/Cas Type I-F Cascade component monomer, 19 kDa RNA
|
| Buffer: |
50 mM HEPES 150 mM NaCl 1mM DTT 1mM EDTA, pH: 7 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2015 Jun 27
|
Modulating the Cascade architecture of a minimal Type I-F CRISPR-Cas system.
Nucleic Acids Res 44(12):5872-82 (2016)
Gleditzsch D, Müller-Esparza H, Pausch P, Sharma K, Dwarakanath S, Urlaub H, Bange G, Randau L
|
| RgGuinier |
5.4 |
nm |
| Dmax |
18.4 |
nm |
|
|