UniProt ID: A4Y6G3 (1-183) Cas6f: CRISPR/Cas Type I-F Cascade component (CRISPR-associated protein, Csy4 family)
UniProt ID: A4Y6G2 (1-336) Cas5fv: CRISPR/Cas Type I-F Cascade component (Uncharacterized protein, Sputcn32_1822)
UniProt ID: A4Y6G1 (1-315) Nonameric Cas7fv: CRISPR/Cas Type I-F Cascade component (Uncharacterized protein, Sputcn32_1821)
UniProt ID: (None-None) long-crRNA: CRISPR/Cas Type I-F Cascade component
|
|
|

|
| Sample: |
Cas6f: CRISPR/Cas Type I-F Cascade component (CRISPR-associated protein, Csy4 family) monomer, 21 kDa Shewanella putrefaciens protein
Cas5fv: CRISPR/Cas Type I-F Cascade component (Uncharacterized protein, Sputcn32_1822) monomer, 38 kDa Shewanella putrefaciens protein
Nonameric Cas7fv: CRISPR/Cas Type I-F Cascade component (Uncharacterized protein, Sputcn32_1821) nonamer, 335 kDa Shewanella putrefaciens protein
Long-crRNA: CRISPR/Cas Type I-F Cascade component monomer, 25 kDa RNA
|
| Buffer: |
50 mM HEPES 150 mM NaCl 1mM DTT 1mM EDTA, pH: 7 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Jan 30
|
Modulating the Cascade architecture of a minimal Type I-F CRISPR-Cas system.
Nucleic Acids Res 44(12):5872-82 (2016)
Gleditzsch D, Müller-Esparza H, Pausch P, Sharma K, Dwarakanath S, Urlaub H, Bange G, Randau L
|
| RgGuinier |
6.5 |
nm |
| Dmax |
21.6 |
nm |
|
|
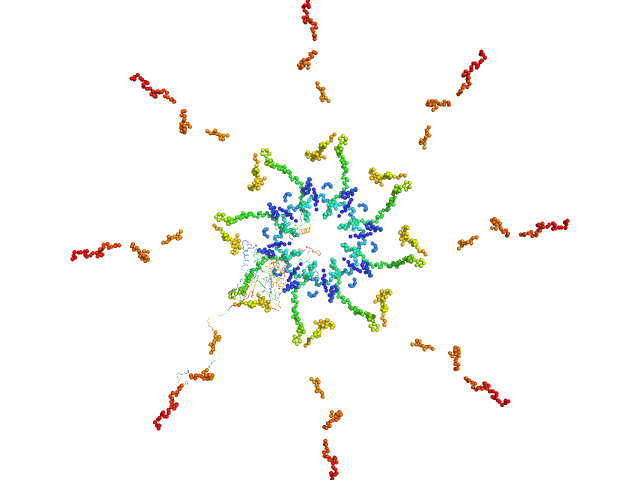
UniProt ID: Q9ZT82 (570-1208) Callose synthase
|
|
|
|
| Sample: |
Callose synthase octamer, 633 kDa Arabidopsis thaliana protein
|
| Buffer: |
Tris, 50 mM NaCl, pH: 7.3 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2015 Jun 2
|
Structural Characterization of Cell Wall and Plasma Membrane Proteins of Arabidopsis thaliana
University of Hamburg Dissertation 8022 (2016)
Haifa El Kilani
|
| RgGuinier |
8.0 |
nm |
| Dmax |
30.0 |
nm |
| VolumePorod |
1033 |
nm3 |
|
|
UniProt ID: Q08840 (43-271) Iron-regulated outer membrane lipoprotein FrpD
|
|
|

|
| Sample: |
Iron-regulated outer membrane lipoprotein FrpD monomer, 27 kDa Neisseria meningitidis protein
|
| Buffer: |
10 mM Tris-HCl 150 mM NaCl 0.01% NaN3, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 Oct 19
|
Structural basis of the interaction between the putative adhesion-involved and iron-regulated FrpD and FrpC proteins of Neisseria meningitidis.
Sci Rep 7:40408 (2017)
Sviridova E, Rezacova P, Bondar A, Veverka V, Novak P, Schenk G, Svergun DI, Kuta Smatanova I, Bumba L
|
| RgGuinier |
2.2 |
nm |
| Dmax |
6.5 |
nm |
| VolumePorod |
41 |
nm3 |
|
|
UniProt ID: Q08840 (43-271) Iron-regulated outer membrane lipoprotein FrpD
UniProt ID: Q9JYV5 (1-414) Iron-regulated protein FrpC
|
|
|

|
| Sample: |
Iron-regulated outer membrane lipoprotein FrpD monomer, 27 kDa Neisseria meningitidis protein
Iron-regulated protein FrpC monomer, 46 kDa Neisseria meningitidis protein
|
| Buffer: |
50 mM Tris-HCl 150 mM NaCl 0.01% NaN3, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 Oct 19
|
Structural basis of the interaction between the putative adhesion-involved and iron-regulated FrpD and FrpC proteins of Neisseria meningitidis.
Sci Rep 7:40408 (2017)
Sviridova E, Rezacova P, Bondar A, Veverka V, Novak P, Schenk G, Svergun DI, Kuta Smatanova I, Bumba L
|
| RgGuinier |
3.7 |
nm |
| Dmax |
13.5 |
nm |
| VolumePorod |
123 |
nm3 |
|
|
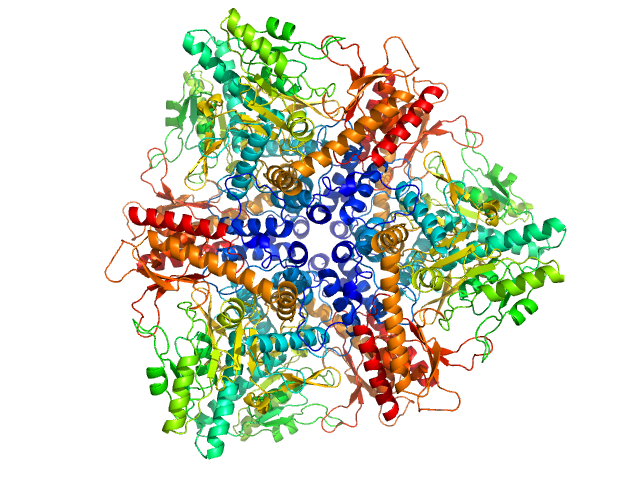
UniProt ID: P69908 (None-None) Glutamate decarboxylase alpha (GadA) from E. coli
|
|
|
|
| Sample: |
Glutamate decarboxylase alpha (GadA) from E. coli monomer, 53 kDa Escherichia coli protein
|
| Buffer: |
50 mM Tris, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2015 Jul 29
|
X-Ray Solution Scattering Study of Four Escherichia coli Enzymes Involved in Stationary-Phase Metabolism.
PLoS One 11(5):e0156105 (2016)
Dadinova LA, Shtykova EV, Konarev PV, Rodina EV, Snalina NE, Vorobyeva NN, Kurilova SA, Nazarova TI, Jeffries CM, Svergun DI
|
| RgGuinier |
4.4 |
nm |
| VolumePorod |
450 |
nm3 |
|
|
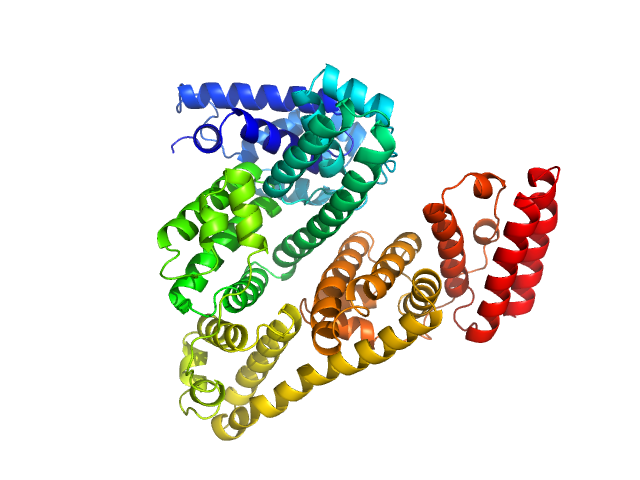
UniProt ID: P02769 (25-607) Bovine serum albumin, monomer
|
|
|
|
| Sample: |
Bovine serum albumin, monomer monomer, 66 kDa Bos taurus protein
|
| Buffer: |
20 mM Tris 142 mM NaCl 5 % Glycerol 1mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Feb 22
|
Bovine Serum Albumin measured by SEC-SAXS
Martha Brennich
|
| RgGuinier |
2.7 |
nm |
| Dmax |
8.2 |
nm |
| VolumePorod |
118 |
nm3 |
|
|
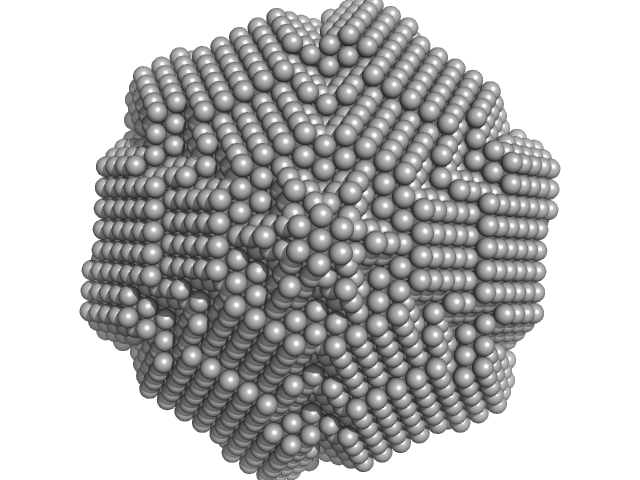
UniProt ID: P21010 (323-785) Primase D5 protein fragment containing the D5N and helicase domain
|
|
|
|
| Sample: |
Primase D5 protein fragment containing the D5N and helicase domain hexamer, 321 kDa Vaccinia virus protein
|
| Buffer: |
20 mM Tris 150 mM NaCL 5% glycerol 1mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2014 Sep 26
|
Domain Organization of Vaccinia Virus Helicase-Primase D5.
J Virol 90(9):4604-4613 (2016)
Hutin S, Ling WL, Round A, Effantin G, Reich S, Iseni F, Tarbouriech N, Schoehn G, Burmeister WP
|
| RgGuinier |
4.8 |
nm |
| Dmax |
14.5 |
nm |
| VolumePorod |
570 |
nm3 |
|
|
UniProt ID: O57173 (1-202) MVA F1L antiapoptotic Bcl-2 viral protein
|
|
|
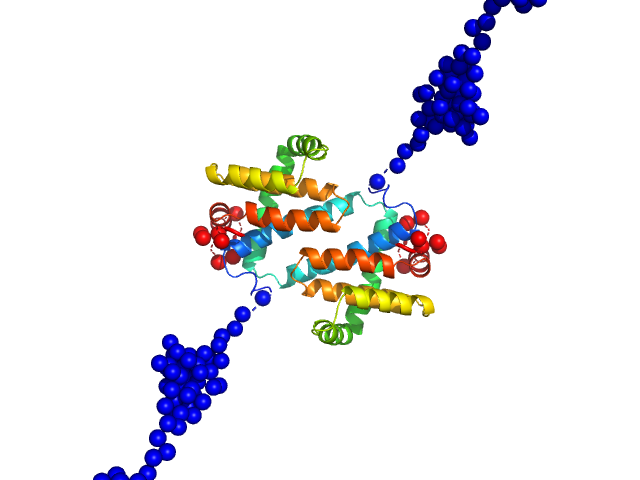
|
| Sample: |
MVA F1L antiapoptotic Bcl-2 viral protein dimer, 51 kDa Vaccinia virus protein
|
| Buffer: |
25 mM HEPES 150 mM NaCl 5 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2016 Apr 15
|
The N Terminus of the Vaccinia Virus Protein F1L Is an Intrinsically Unstructured Region That Is Not Involved in Apoptosis Regulation.
J Biol Chem 291(28):14600-8 (2016)
Caria S, Marshall B, Burton RL, Campbell S, Pantaki-Eimany D, Hawkins CJ, Barry M, Kvansakul M
|
| RgGuinier |
3.4 |
nm |
| Dmax |
16.2 |
nm |
| VolumePorod |
74 |
nm3 |
|
|
UniProt ID: V9P0A9 (None-None) Bacterial chalcone isomerase
|
|
|
|
| Sample: |
Bacterial chalcone isomerase hexamer, 194 kDa Eubacterium ramulus protein
|
| Buffer: |
50 mM sodium phosphate, pH: 6.8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Sep 23
|
Structure and catalytic mechanism of the evolutionarily unique bacterial chalcone isomerase.
Acta Crystallogr D Biol Crystallogr 71(Pt 4):907-17 (2015)
Thomsen M, Tuukkanen A, Dickerhoff J, Palm GJ, Kratzat H, Svergun DI, Weisz K, Bornscheuer UT, Hinrichs W
|
| RgGuinier |
4.0 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
320 |
nm3 |
|
|
UniProt ID: J7I2T6 (None-None) Perivitellin ovorubin-1
UniProt ID: J7HZ90 (None-None) Perivitellin ovorubin-2
UniProt ID: J7I5Z5 (None-None) Perivitellin ovorubin-3
|
|
|

|
| Sample: |
Perivitellin ovorubin-1, 22 kDa Pomacea canaliculata protein
Perivitellin ovorubin-2, 24 kDa Pomacea canaliculata protein
Perivitellin ovorubin-3, 35 kDa Pomacea canaliculata protein
|
| Buffer: |
20 mM Tris-HCl, pH: 8.5 |
| Experiment: |
SAXS
data collected at SAXS2 Beamline, Brazilian Synchrotron Light Laboratory on 2014 Jun 10
|
Apple Snail Perivitellin Precursor Properties Help Explain Predators' Feeding Behavior.
Physiol Biochem Zool 90(4):461-470 (2017)
Cadierno MP, Dreon MS, Heras H
|
| RgGuinier |
4.3 |
nm |
| Dmax |
14.9 |
nm |
| VolumePorod |
526 |
nm3 |
|
|