UniProt ID: Q6ZSD7 (1-400) cDNA FLJ45612 fis, clone BRTHA3025073, highly similar to Actin cross-linking family protein 7 Y259D mutant
|
|
|
|
| Sample: |
CDNA FLJ45612 fis, clone BRTHA3025073, highly similar to Actin cross-linking family protein 7 Y259D mutant monomer, 46 kDa Homo sapiens protein
|
| Buffer: |
PBS, pH: 7.4 |
| Experiment: |
SAXS
data collected at 12-ID-B SAXS/WAXS, Advanced Photon Source (APS), Argonne National Laboratory on 2014 Nov 7
|
In vivo epidermal migration requires focal adhesion targeting of ACF7.
Nat Commun 7:11692 (2016)
Yue J, Zhang Y, Liang WG, Gou X, Lee P, Liu H, Lyu W, Tang WJ, Chen SY, Yang F, Liang H, Wu X
|
| RgGuinier |
3.4 |
nm |
| Dmax |
13.5 |
nm |
| VolumePorod |
55 |
nm3 |
|
|
UniProt ID: T2B9R0 (1-164) Middle East Respiratory Syndrome (MERS) coronavirus nucleocapsid N-Protein (N-terminal domain 1-164)
|
|
|

|
| Sample: |
Middle East Respiratory Syndrome (MERS) coronavirus nucleocapsid N-Protein (N-terminal domain 1-164) monomer, 18 kDa Middle East respiratory … protein
|
| Buffer: |
10 mM HEPES 300 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2015 Oct 5
|
Structural characterization of the N-terminal part of the MERS-CoV nucleocapsid by X-ray diffraction and small-angle X-ray scattering.
Acta Crystallogr D Struct Biol 72(Pt 2):192-202 (2016)
Papageorgiou N, Lichière J, Baklouti A, Ferron F, Sévajol M, Canard B, Coutard B
|
| RgGuinier |
2.0 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
28 |
nm3 |
|
|
UniProt ID: O76064 (345-485) E3 ubiquitin-protein ligase RNF8
UniProt ID: P61088 (1-152) Ubiquitin-conjugating enzyme E2 N double mutant (C87K, K92A)
UniProt ID: P0CG48 (1-76) Polyubiquitin-C
|
|
|

|
| Sample: |
E3 ubiquitin-protein ligase RNF8 dimer, 35 kDa Homo sapiens protein
Ubiquitin-conjugating enzyme E2 N double mutant (C87K, K92A) dimer, 36 kDa Homo sapiens protein
Polyubiquitin-C dimer, 17 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES 200 mM NaCl 0.01 mM ZnSO4 1 mM DTT, pH: 6.8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2015 Sep 8
|
RNF8 E3 Ubiquitin Ligase Stimulates Ubc13 E2 Conjugating Activity That Is Essential for DNA Double Strand Break Signaling and BRCA1 Tumor Suppressor Recruitment.
J Biol Chem 291(18):9396-410 (2016)
Hodge CD, Ismail IH, Edwards RA, Hura GL, Xiao AT, Tainer JA, Hendzel MJ, Glover JN
|
| RgGuinier |
4.5 |
nm |
| Dmax |
18.9 |
nm |
| VolumePorod |
119 |
nm3 |
|
|
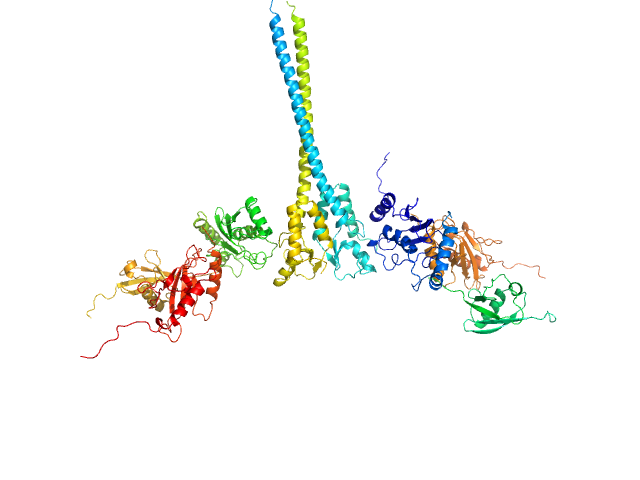
UniProt ID: O76064 (345-485) E3 ubiquitin-protein ligase RNF8
UniProt ID: P61088 (1-152) Ubiquitin-conjugating enzyme E2 N double mutant (C87K, K92A)
UniProt ID: P0CG48 (1-76) Polyubiquitin-C
UniProt ID: Q15819 (1-145) Ubiquitin-conjugating enzyme E2 variant 2
|
|
|
|
| Sample: |
E3 ubiquitin-protein ligase RNF8 dimer, 35 kDa Homo sapiens protein
Ubiquitin-conjugating enzyme E2 N double mutant (C87K, K92A) dimer, 36 kDa Homo sapiens protein
Polyubiquitin-C dimer, 17 kDa Homo sapiens protein
Ubiquitin-conjugating enzyme E2 variant 2 dimer, 34 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES 200 mM NaCl 0.01 mM ZnSO4 1 mM DTT, pH: 6.8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2015 Sep 8
|
RNF8 E3 Ubiquitin Ligase Stimulates Ubc13 E2 Conjugating Activity That Is Essential for DNA Double Strand Break Signaling and BRCA1 Tumor Suppressor Recruitment.
J Biol Chem 291(18):9396-410 (2016)
Hodge CD, Ismail IH, Edwards RA, Hura GL, Xiao AT, Tainer JA, Hendzel MJ, Glover JN
|
| RgGuinier |
5.4 |
nm |
| Dmax |
23.8 |
nm |
| VolumePorod |
214 |
nm3 |
|
|
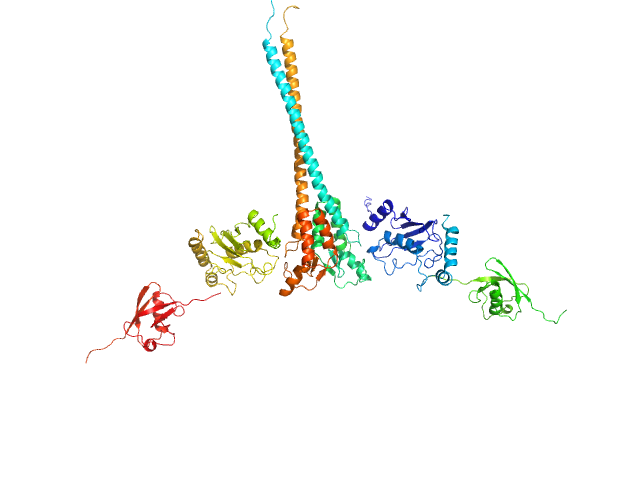
UniProt ID: P61088 (1-152) Ubiquitin-conjugating enzyme E2 N double mutant (C87K, K92A)
UniProt ID: P0CG48 (1-76) Polyubiquitin-C
UniProt ID: O76064 (345-485) E3 ubiquitin-protein ligase RNF8 mutant (L451D)
|
|
|
|
| Sample: |
Ubiquitin-conjugating enzyme E2 N double mutant (C87K, K92A) dimer, 36 kDa Homo sapiens protein
Polyubiquitin-C dimer, 17 kDa Homo sapiens protein
E3 ubiquitin-protein ligase RNF8 mutant (L451D) dimer, 35 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES 200 mM NaCl 0.01 mM ZnSO4 1 mM DTT, pH: 6.8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2015 Sep 8
|
RNF8 E3 Ubiquitin Ligase Stimulates Ubc13 E2 Conjugating Activity That Is Essential for DNA Double Strand Break Signaling and BRCA1 Tumor Suppressor Recruitment.
J Biol Chem 291(18):9396-410 (2016)
Hodge CD, Ismail IH, Edwards RA, Hura GL, Xiao AT, Tainer JA, Hendzel MJ, Glover JN
|
| RgGuinier |
4.3 |
nm |
| Dmax |
18.9 |
nm |
| VolumePorod |
111 |
nm3 |
|
|
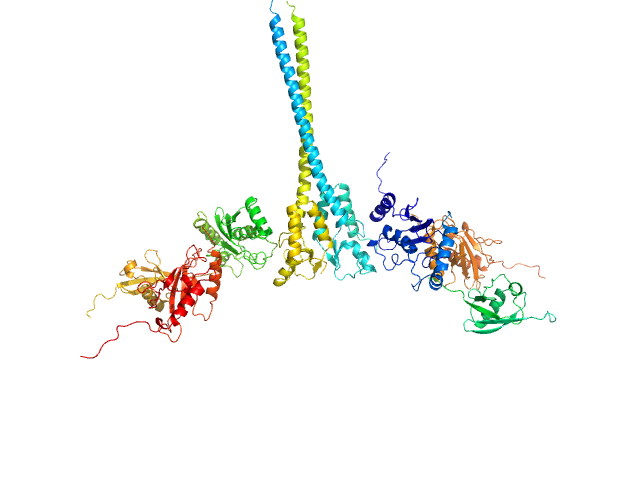
UniProt ID: P61088 (1-152) Ubiquitin-conjugating enzyme E2 N double mutant (C87K, K92A)
UniProt ID: P0CG48 (1-76) Polyubiquitin-C
UniProt ID: Q15819 (1-145) Ubiquitin-conjugating enzyme E2 variant 2
UniProt ID: O76064 (345-485) E3 ubiquitin-protein ligase RNF8 mutant (L451D)
|
|
|
|
| Sample: |
Ubiquitin-conjugating enzyme E2 N double mutant (C87K, K92A) dimer, 36 kDa Homo sapiens protein
Polyubiquitin-C dimer, 17 kDa Homo sapiens protein
Ubiquitin-conjugating enzyme E2 variant 2 dimer, 34 kDa Homo sapiens protein
E3 ubiquitin-protein ligase RNF8 mutant (L451D) dimer, 35 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES 200 mM NaCl 0.01 mM ZnSO4 1 mM DTT, pH: 6.8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2015 Sep 8
|
RNF8 E3 Ubiquitin Ligase Stimulates Ubc13 E2 Conjugating Activity That Is Essential for DNA Double Strand Break Signaling and BRCA1 Tumor Suppressor Recruitment.
J Biol Chem 291(18):9396-410 (2016)
Hodge CD, Ismail IH, Edwards RA, Hura GL, Xiao AT, Tainer JA, Hendzel MJ, Glover JN
|
| RgGuinier |
5.2 |
nm |
| Dmax |
23.8 |
nm |
| VolumePorod |
192 |
nm3 |
|
|
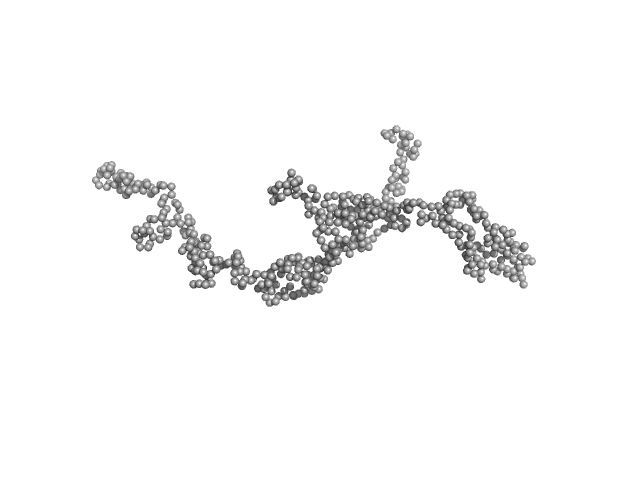
UniProt ID: P11532 (1991-2694) Dystrophin central domain repeats 16 to 21 (Δ2146-2305; Becker muscular dystrophy variant, deletion of exons 45-47)
|
|
|
|
| Sample: |
Dystrophin central domain repeats 16 to 21 (Δ2146-2305; Becker muscular dystrophy variant, deletion of exons 45-47) monomer, 64 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris 150 mM NaCl 1 mM EDTA 2% glycerol 5% acetonitrile, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2014 Feb 5
|
Dystrophin's central domain forms a complex filament that becomes disorganized by in-frame deletions.
J Biol Chem 293(18):6637-6646 (2018)
Delalande O, Molza AE, Dos Santos Morais R, Chéron A, Pollet É, Raguenes-Nicol C, Tascon C, Giudice E, Guilbaud M, Nicolas A, Bondon A, Leturcq F, Férey N, Baaden M, Perez J, Roblin P, Piétri-Rouxel F, Hubert JF, Czjzek M, Le Rumeur E
|
| RgGuinier |
6.0 |
nm |
| Dmax |
21.0 |
nm |
| VolumePorod |
184 |
nm3 |
|
|
UniProt ID: O43852 (68-315) Human Calumenin
|
|
|
|
| Sample: |
Human Calumenin monomer, 29 kDa Homo sapiens protein
|
| Buffer: |
25 mM Na-HEPES, 25 mM NaCl, 2.5 mM CaCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2016 Feb 12
|
Ca-Dependent Folding of Human Calumenin.
PLoS One 11(3):e0151547 (2016)
Mazzorana M, Hussain R, Sorensen T
|
| RgGuinier |
2.3 |
nm |
| Dmax |
6.5 |
nm |
| VolumePorod |
49 |
nm3 |
|
|
UniProt ID: A4H5F0 (1-547) Stress-induced protein sti1
|
|
|
|
| Sample: |
Stress-induced protein sti1 monomer, 62 kDa Leishmania braziliensis protein
|
| Buffer: |
25 mM Tris 100 mM NaCl 1 mM EDTA 1 mM β-mercaptoethanol, pH: 7.5 |
| Experiment: |
SAXS
data collected at SAXS1 Beamline, Brazilian Synchrotron Light Laboratory on 2016 Feb 21
|
Low sequence identity but high structural and functional conservation: The case of Hsp70/Hsp90 organizing protein (Hop/Sti1) of Leishmania braziliensis.
Arch Biochem Biophys 600:12-22 (2016)
Batista FAH, Seraphim TV, Santos CA, Gonzaga MR, Barbosa LRS, Ramos CHI, Borges JC
|
| RgGuinier |
4.5 |
nm |
| Dmax |
18.0 |
nm |
| VolumePorod |
94 |
nm3 |
|
|
UniProt ID: A4H5F0 (171-547) Stress-induced protein sti1 (Hop TPR2A-TPR2B-DP2 construct)
|
|
|

|
| Sample: |
Stress-induced protein sti1 (Hop TPR2A-TPR2B-DP2 construct) monomer, 44 kDa Leishmania braziliensis protein
|
| Buffer: |
25 mM Tris 100 mM NaCl 1 mM EDTA 1 mM β-mercaptoethanol, pH: 7.5 |
| Experiment: |
SAXS
data collected at SAXS2 Beamline, Brazilian Synchrotron Light Laboratory on 2016 Feb 21
|
Low sequence identity but high structural and functional conservation: The case of Hsp70/Hsp90 organizing protein (Hop/Sti1) of Leishmania braziliensis.
Arch Biochem Biophys 600:12-22 (2016)
Batista FAH, Seraphim TV, Santos CA, Gonzaga MR, Barbosa LRS, Ramos CHI, Borges JC
|
| RgGuinier |
3.8 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
65 |
nm3 |
|
|