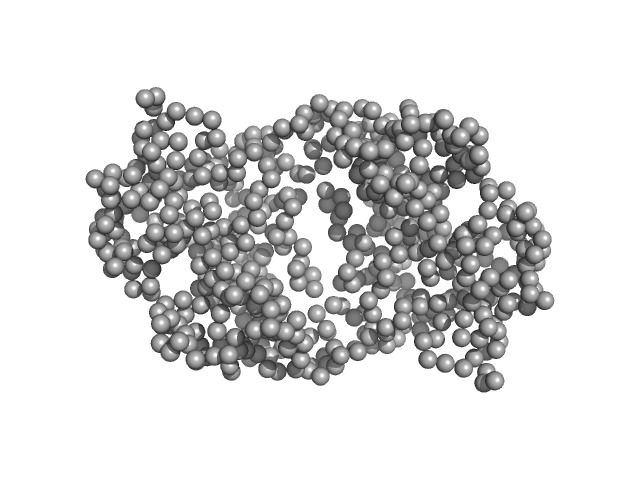
UniProt ID: Q8TB36 (1-318) Ganglioside-induced differentiation-associated protein 1, construct GDAP1∆319-358
|
|
|
|
| Sample: |
Ganglioside-induced differentiation-associated protein 1, construct GDAP1∆319-358 dimer, 74 kDa Homo sapiens protein
|
| Buffer: |
25 mM HEPES, 300 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Jul 11
|
Structure of the Complete Dimeric Human GDAP1 Core Domain Provides Insights into Ligand Binding and Clustering of Disease Mutations
Frontiers in Molecular Biosciences 7 (2021)
Nguyen G, Sutinen A, Raasakka A, Muruganandam G, Loris R, Kursula P
|
| RgGuinier |
3.1 |
nm |
| Dmax |
9.6 |
nm |
| VolumePorod |
121 |
nm3 |
|
|
UniProt ID: Q8TB36 (1-318) Ganglioside-induced differentiation-associated protein 1, construct GDAP1∆319-358
|
|
|

|
| Sample: |
Ganglioside-induced differentiation-associated protein 1, construct GDAP1∆319-358 monomer, 37 kDa Homo sapiens protein
|
| Buffer: |
25 mM HEPES, 300 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Jul 11
|
Structure of the Complete Dimeric Human GDAP1 Core Domain Provides Insights into Ligand Binding and Clustering of Disease Mutations
Frontiers in Molecular Biosciences 7 (2021)
Nguyen G, Sutinen A, Raasakka A, Muruganandam G, Loris R, Kursula P
|
| RgGuinier |
2.7 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
69 |
nm3 |
|
|
UniProt ID: Q8TB36 (1-318) Ganglioside-induced differentiation-associated protein 1, construct GDAP1∆319-358
|
|
|

|
| Sample: |
Ganglioside-induced differentiation-associated protein 1, construct GDAP1∆319-358 monomer, 37 kDa Homo sapiens protein
|
| Buffer: |
25 mM HEPES, 300 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Jul 11
|
Structure of the Complete Dimeric Human GDAP1 Core Domain Provides Insights into Ligand Binding and Clustering of Disease Mutations
Frontiers in Molecular Biosciences 7 (2021)
Nguyen G, Sutinen A, Raasakka A, Muruganandam G, Loris R, Kursula P
|
| RgGuinier |
2.5 |
nm |
| Dmax |
8.6 |
nm |
| VolumePorod |
65 |
nm3 |
|
|
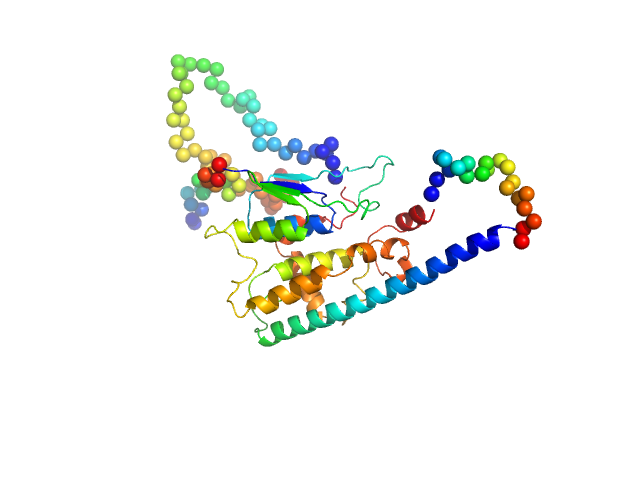
UniProt ID: Q96MZ0 (None-None) Ganglioside-induced differentiation-associated protein 1-like 1 (∆125-143 isoform)
|
|
|
|
| Sample: |
Ganglioside-induced differentiation-associated protein 1-like 1 (∆125-143 isoform) monomer, 44 kDa Homo sapiens protein
|
| Buffer: |
20 mM TRIS pH 7.5, 150 mM NaCl, 1 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 Dec 10
|
Structure of the Complete Dimeric Human GDAP1 Core Domain Provides Insights into Ligand Binding and Clustering of Disease Mutations
Frontiers in Molecular Biosciences 7 (2021)
Nguyen G, Sutinen A, Raasakka A, Muruganandam G, Loris R, Kursula P
|
| RgGuinier |
2.7 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
73 |
nm3 |
|
|
UniProt ID: Q5W264 (1-189) 4-hydroxy-2,2'-bipyrrole-5-methanol synthase PigH
|
|
|
|
| Sample: |
4-hydroxy-2,2'-bipyrrole-5-methanol synthase PigH monomer, 22 kDa Serratia sp. protein
|
| Buffer: |
25 mM Tris, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2020 Mar 3
|
Solution Structure and Conformational Dynamics of a Doublet Acyl Carrier Protein from Prodigiosin Biosynthesis.
Biochemistry (2021)
Thongkawphueak T, Winter AJ, Williams C, Maple HJ, Soontaranon S, Kaewhan C, Campopiano DJ, Crump MP, Wattana-Amorn P
|
| RgGuinier |
2.9 |
nm |
| Dmax |
9.9 |
nm |
| VolumePorod |
83 |
nm3 |
|
|
UniProt ID: Q5W264 (1-189) 4-hydroxy-2,2'-bipyrrole-5-methanol synthase PigH
|
|
|
|
| Sample: |
4-hydroxy-2,2'-bipyrrole-5-methanol synthase PigH monomer, 22 kDa Serratia sp. protein
|
| Buffer: |
25 mM Tris, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2020 Mar 3
|
Solution Structure and Conformational Dynamics of a Doublet Acyl Carrier Protein from Prodigiosin Biosynthesis.
Biochemistry (2021)
Thongkawphueak T, Winter AJ, Williams C, Maple HJ, Soontaranon S, Kaewhan C, Campopiano DJ, Crump MP, Wattana-Amorn P
|
| RgGuinier |
2.8 |
nm |
| Dmax |
9.7 |
nm |
| VolumePorod |
72 |
nm3 |
|
|
UniProt ID: P42196 (1-239) Sensory rhodopsin II from Natronbacterium pharaonis
UniProt ID: P42259 (3-534) Sensory rhodopsin II transducer from Natronomonas pharaonis
|
|
|

|
| Sample: |
Sensory rhodopsin II from Natronbacterium pharaonis dimer, 53 kDa Natronomonas pharaonis protein
Sensory rhodopsin II transducer from Natronomonas pharaonis dimer, 116 kDa Natronomonas pharaonis protein
|
| Buffer: |
150 mM NaCl, 25 mM Na/Na-Pi, 1.0 mM EDTA, 0.05% DDM, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Dec 8
|
Ambiguities in and completeness of SAS data analysis of membrane proteins: the case of the sensory rhodopsin II–transducer complex
Acta Crystallographica Section D Structural Biology 77(11) (2021)
Ryzhykau Y, Vlasov A, Orekhov P, Rulev M, Rogachev A, Vlasova A, Kazantsev A, Verteletskiy D, Skoi V, Brennich M, Pernot P, Murugova T, Gordeliy V, Kuklin A
|
| RgGuinier |
7.6 |
nm |
| Dmax |
41.0 |
nm |
|
|
UniProt ID: P42196 (1-239) Sensory rhodopsin II from Natronbacterium pharaonis
UniProt ID: P42259 (3-534) Sensory rhodopsin II transducer from Natronomonas pharaonis
|
|
|

|
| Sample: |
Sensory rhodopsin II from Natronbacterium pharaonis dimer, 53 kDa Natronomonas pharaonis protein
Sensory rhodopsin II transducer from Natronomonas pharaonis dimer, 116 kDa Natronomonas pharaonis protein
|
| Buffer: |
150 mM NaCl, 25 mM Na/Na-Pi, 1.0 mM EDTA, 0.05% DDM (D2O buffer), pH: 8 |
| Experiment: |
SANS
data collected at YuMO SANS TOF spectrometer, IBR-2, Frank Laboratory of Neutron Physics, Joint Institute for Nuclear Research on 2019 Jan 25
|
Ambiguities in and completeness of SAS data analysis of membrane proteins: the case of the sensory rhodopsin II–transducer complex
Acta Crystallographica Section D Structural Biology 77(11) (2021)
Ryzhykau Y, Vlasov A, Orekhov P, Rulev M, Rogachev A, Vlasova A, Kazantsev A, Verteletskiy D, Skoi V, Brennich M, Pernot P, Murugova T, Gordeliy V, Kuklin A
|
| RgGuinier |
8.9 |
nm |
| Dmax |
39.0 |
nm |
|
|
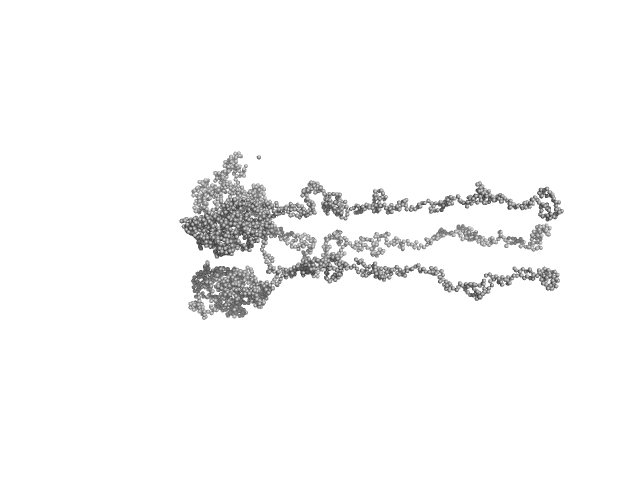
UniProt ID: P42196 (1-239) Sensory rhodopsin II from Natronbacterium pharaonis
UniProt ID: P42259 (3-534) Sensory rhodopsin II transducer from Natronomonas pharaonis
|
|
|
|
| Sample: |
Sensory rhodopsin II from Natronbacterium pharaonis dimer, 53 kDa Natronomonas pharaonis protein
Sensory rhodopsin II transducer from Natronomonas pharaonis dimer, 116 kDa Natronomonas pharaonis protein
|
| Buffer: |
1400 mM NaCl, 49.4 mM Na/Na-Pi, 1.0 mM EDTA, 0.05% DDM (D2O buffer), pH: 8 |
| Experiment: |
SANS
data collected at YuMO SANS TOF spectrometer, IBR-2, Frank Laboratory of Neutron Physics, Joint Institute for Nuclear Research on 2019 Feb 10
|
Ambiguities in and completeness of SAS data analysis of membrane proteins: the case of the sensory rhodopsin II–transducer complex
Acta Crystallographica Section D Structural Biology 77(11) (2021)
Ryzhykau Y, Vlasov A, Orekhov P, Rulev M, Rogachev A, Vlasova A, Kazantsev A, Verteletskiy D, Skoi V, Brennich M, Pernot P, Murugova T, Gordeliy V, Kuklin A
|
| RgGuinier |
8.6 |
nm |
| Dmax |
39.0 |
nm |
|
|
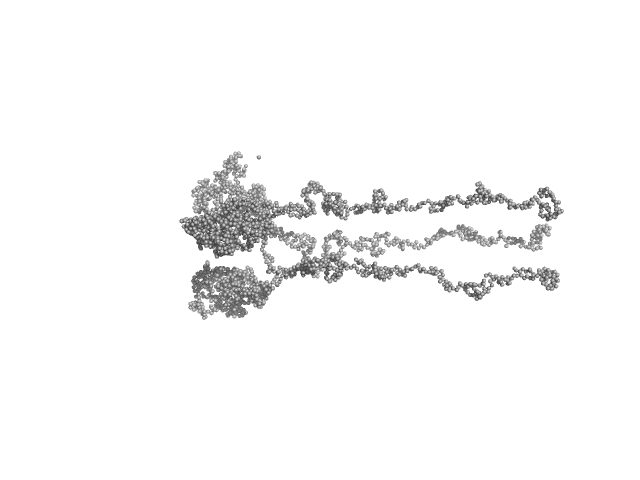
UniProt ID: P42196 (1-239) Sensory rhodopsin II from Natronbacterium pharaonis
UniProt ID: P42259 (3-534) Sensory rhodopsin II transducer from Natronomonas pharaonis
|
|
|
|
| Sample: |
Sensory rhodopsin II from Natronbacterium pharaonis dimer, 53 kDa Natronomonas pharaonis protein
Sensory rhodopsin II transducer from Natronomonas pharaonis dimer, 116 kDa Natronomonas pharaonis protein
|
| Buffer: |
2800 mM NaCl, 76.6 mM Na/Na-Pi, 1.0 mM EDTA, 0.05% DDM (D2O buffer), pH: 8 |
| Experiment: |
SANS
data collected at YuMO SANS TOF spectrometer, IBR-2, Frank Laboratory of Neutron Physics, Joint Institute for Nuclear Research on 2019 Feb 10
|
Ambiguities in and completeness of SAS data analysis of membrane proteins: the case of the sensory rhodopsin II–transducer complex
Acta Crystallographica Section D Structural Biology 77(11) (2021)
Ryzhykau Y, Vlasov A, Orekhov P, Rulev M, Rogachev A, Vlasova A, Kazantsev A, Verteletskiy D, Skoi V, Brennich M, Pernot P, Murugova T, Gordeliy V, Kuklin A
|
| RgGuinier |
9.0 |
nm |
| Dmax |
39.0 |
nm |
|
|