UniProt ID: P42196 (1-239) Sensory rhodopsin II from Natronbacterium pharaonis
UniProt ID: P42259 (3-534) Sensory rhodopsin II transducer from Natronomonas pharaonis
|
|
|

|
| Sample: |
Sensory rhodopsin II from Natronbacterium pharaonis dimer, 53 kDa Natronomonas pharaonis protein
Sensory rhodopsin II transducer from Natronomonas pharaonis dimer, 116 kDa Natronomonas pharaonis protein
|
| Buffer: |
4000 mM NaCl, 100 mM Na/Na-Pi, 1.0 mM EDTA, 0.05% DDM (D2O buffer), pH: 8 |
| Experiment: |
SANS
data collected at YuMO SANS TOF spectrometer, IBR-2, Frank Laboratory of Neutron Physics, Joint Institute for Nuclear Research on 2019 Jan 25
|
Ambiguities in and completeness of SAS data analysis of membrane proteins: the case of the sensory rhodopsin II–transducer complex
Acta Crystallographica Section D Structural Biology 77(11) (2021)
Ryzhykau Y, Vlasov A, Orekhov P, Rulev M, Rogachev A, Vlasova A, Kazantsev A, Verteletskiy D, Skoi V, Brennich M, Pernot P, Murugova T, Gordeliy V, Kuklin A
|
| RgGuinier |
10.4 |
nm |
| Dmax |
39.0 |
nm |
|
|
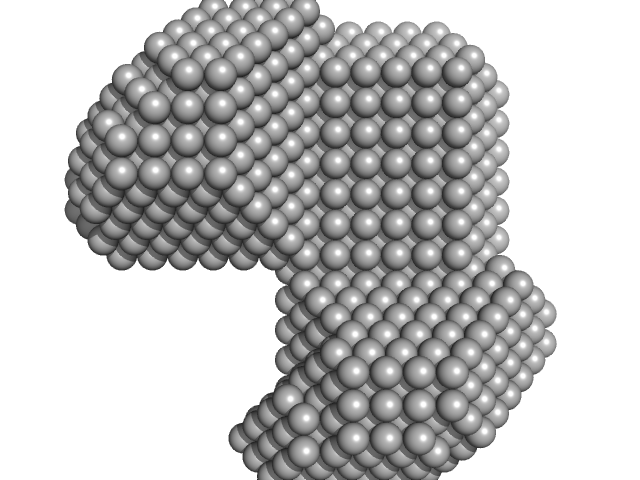
UniProt ID: P50481 (148-227) LIM/homeobox protein Lhx3
|
|
|
|
| Sample: |
LIM/homeobox protein Lhx3 monomer, 10 kDa Mus musculus protein
|
| Buffer: |
20 mM sodium phosphate monobasic/dibasic, 100 mM NaCl, 1 mM DTT, pH: 7.4 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2018 Nov 2
|
Contrasting DNA-binding behaviour by ISL1 and LHX3 underpins differential gene targeting in neuronal cell specification
Journal of Structural Biology: X :100043 (2020)
Smith N, Wilkinson-White L, Kwan A, Trewhella J, Matthews J
|
| RgGuinier |
2.1 |
nm |
| Dmax |
7.0 |
nm |
| VolumePorod |
18 |
nm3 |
|
|
UniProt ID: P50481 (28-227) LIM/homeobox protein Lhx3
UniProt ID: P61372 (262-291) Insulin gene enhancer protein ISL-1
|
|
|
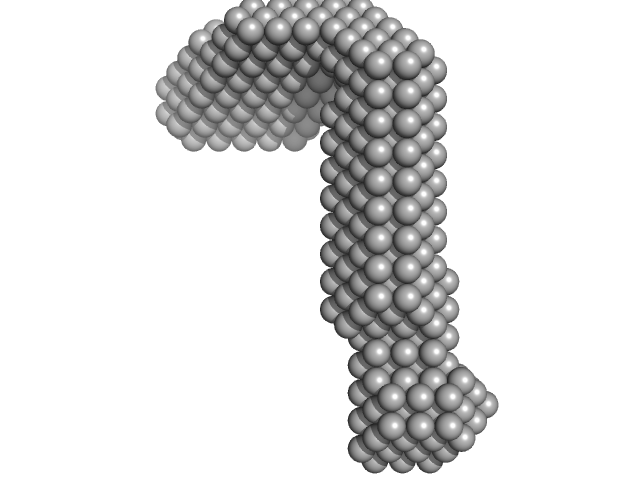
|
| Sample: |
LIM/homeobox protein Lhx3 monomer, 23 kDa Mus musculus protein
Insulin gene enhancer protein ISL-1 monomer, 4 kDa Mus musculus protein
|
| Buffer: |
20 mM sodium phosphate monobasic/dibasic, 100 mM NaCl, 1 mM DTT, pH: 7.4 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2018 Nov 2
|
Contrasting DNA-binding behaviour by ISL1 and LHX3 underpins differential gene targeting in neuronal cell specification
Journal of Structural Biology: X :100043 (2020)
Smith N, Wilkinson-White L, Kwan A, Trewhella J, Matthews J
|
| RgGuinier |
3.3 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
39 |
nm3 |
|
|
UniProt ID: P61372 (164-261) Insulin gene enhancer protein ISL-1
UniProt ID: P50481 (148-227) LIM/homeobox protein Lhx3
|
|
|
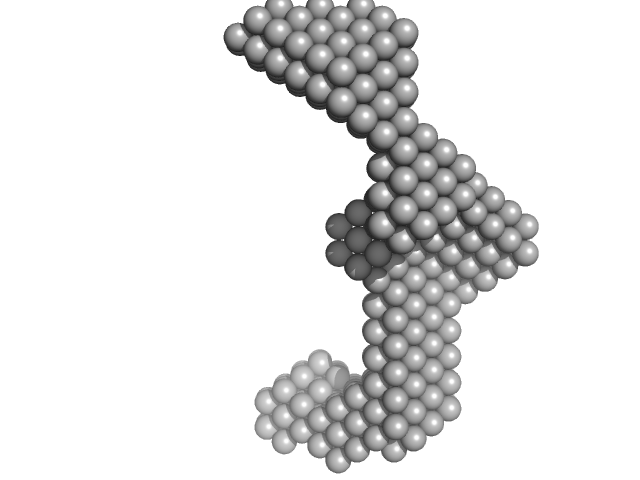
|
| Sample: |
Insulin gene enhancer protein ISL-1 monomer, 12 kDa Mus musculus protein
LIM/homeobox protein Lhx3 monomer, 9 kDa Mus musculus protein
|
| Buffer: |
20 mM sodium phosphate monobasic/dibasic, 100 mM NaCl, 1 mM DTT, pH: 7.4 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2018 Nov 2
|
Contrasting DNA-binding behaviour by ISL1 and LHX3 underpins differential gene targeting in neuronal cell specification
Journal of Structural Biology: X :100043 (2020)
Smith N, Wilkinson-White L, Kwan A, Trewhella J, Matthews J
|
| RgGuinier |
3.4 |
nm |
| Dmax |
12.7 |
nm |
| VolumePorod |
40 |
nm3 |
|
|
UniProt ID: P61372 (179-291) Insulin gene enhancer protein ISL-1
UniProt ID: P50481 (28-227) LIM/homeobox protein Lhx3
|
|
|

|
| Sample: |
Insulin gene enhancer protein ISL-1 monomer, 14 kDa Mus musculus protein
LIM/homeobox protein Lhx3 monomer, 23 kDa Mus musculus protein
|
| Buffer: |
20 mM sodium phosphate monobasic/dibasic, 100 mM NaCl, 1 mM DTT, pH: 7.4 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2018 Nov 2
|
Contrasting DNA-binding behaviour by ISL1 and LHX3 underpins differential gene targeting in neuronal cell specification
Journal of Structural Biology: X :100043 (2020)
Smith N, Wilkinson-White L, Kwan A, Trewhella J, Matthews J
|
| RgGuinier |
3.4 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
76 |
nm3 |
|
|
UniProt ID: None (None-None) M100 oligonucleotide
UniProt ID: P50481 (148-227) LIM/homeobox protein Lhx3
|
|
|

|
| Sample: |
M100 oligonucleotide monomer, 12 kDa DNA
LIM/homeobox protein Lhx3 monomer, 10 kDa Mus musculus protein
|
| Buffer: |
20 mM sodium phosphate monobasic/dibasic, 100 mM NaCl, 1 mM DTT, pH: 7.4 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2018 Nov 2
|
Contrasting DNA-binding behaviour by ISL1 and LHX3 underpins differential gene targeting in neuronal cell specification
Journal of Structural Biology: X :100043 (2020)
Smith N, Wilkinson-White L, Kwan A, Trewhella J, Matthews J
|
| RgGuinier |
2.1 |
nm |
| Dmax |
7.0 |
nm |
| VolumePorod |
27 |
nm3 |
|
|
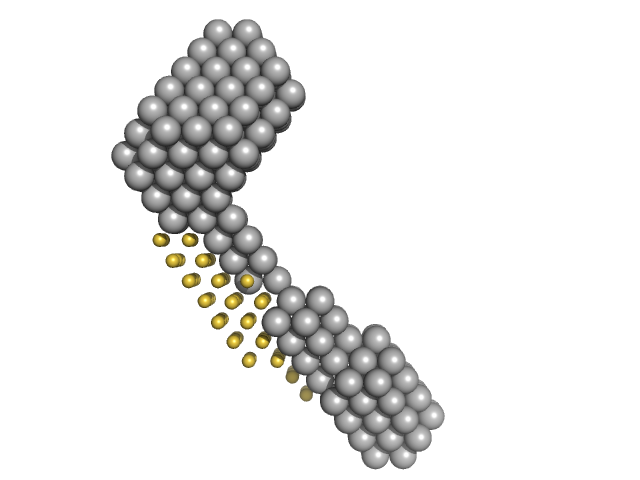
UniProt ID: None (None-None) M100 oligonucleotide
UniProt ID: P50481 (28-227) LIM/homeobox protein Lhx3
UniProt ID: P61372 (262-291) Insulin gene enhancer protein ISL-1
|
|
|
|
| Sample: |
M100 oligonucleotide monomer, 12 kDa DNA
LIM/homeobox protein Lhx3 monomer, 23 kDa Mus musculus protein
Insulin gene enhancer protein ISL-1 monomer, 4 kDa Mus musculus protein
|
| Buffer: |
20 mM sodium phosphate monobasic/dibasic, 100 mM NaCl, 1 mM DTT, pH: 7.4 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2018 Nov 2
|
Contrasting DNA-binding behaviour by ISL1 and LHX3 underpins differential gene targeting in neuronal cell specification
Journal of Structural Biology: X :100043 (2020)
Smith N, Wilkinson-White L, Kwan A, Trewhella J, Matthews J
|
| RgGuinier |
3.6 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
49 |
nm3 |
|
|
UniProt ID: None (None-None) M100 oligonucleotide
UniProt ID: P61372 (164-261) Insulin gene enhancer protein ISL-1
UniProt ID: P50481 (148-227) LIM/homeobox protein Lhx3
|
|
|
|
| Sample: |
M100 oligonucleotide monomer, 12 kDa DNA
Insulin gene enhancer protein ISL-1 monomer, 12 kDa Mus musculus protein
LIM/homeobox protein Lhx3 monomer, 9 kDa Mus musculus protein
|
| Buffer: |
20 mM sodium phosphate monobasic/dibasic, 100 mM NaCl, 1 mM DTT, pH: 7.4 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2018 Nov 2
|
Contrasting DNA-binding behaviour by ISL1 and LHX3 underpins differential gene targeting in neuronal cell specification
Journal of Structural Biology: X :100043 (2020)
Smith N, Wilkinson-White L, Kwan A, Trewhella J, Matthews J
|
| RgGuinier |
2.4 |
nm |
| Dmax |
7.5 |
nm |
| VolumePorod |
41 |
nm3 |
|
|
UniProt ID: None (None-None) M100 oligonucleotide
UniProt ID: P61372 (179-291) Insulin gene enhancer protein ISL-1
UniProt ID: P50481 (28-227) LIM/homeobox protein Lhx3
|
|
|
|
| Sample: |
M100 oligonucleotide monomer, 12 kDa DNA
Insulin gene enhancer protein ISL-1 monomer, 14 kDa Mus musculus protein
LIM/homeobox protein Lhx3 monomer, 23 kDa Mus musculus protein
|
| Buffer: |
20 mM sodium phosphate monobasic/dibasic, 100 mM NaCl, 1 mM DTT, pH: 7.4 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2018 Nov 2
|
Contrasting DNA-binding behaviour by ISL1 and LHX3 underpins differential gene targeting in neuronal cell specification
Journal of Structural Biology: X :100043 (2020)
Smith N, Wilkinson-White L, Kwan A, Trewhella J, Matthews J
|
| RgGuinier |
3.6 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
70 |
nm3 |
|
|
UniProt ID: P07602 (60-140) Prosaposin (Saposin A)
|
|
|
|
| Sample: |
Prosaposin (Saposin A) monomer, 10 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris-HCl, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12-ID-B SAXS/WAXS, Advanced Photon Source (APS), Argonne National Laboratory on 2020 Feb 18
|
Stable Picodisc Assemblies from Saposin Proteins and Branched Detergents.
Biochemistry 60(14):1108-1119 (2021)
Kurgan KW, Chen B, Brown KA, Falco Cobra P, Ye X, Ge Y, Gellman SH
|
| RgGuinier |
1.5 |
nm |
| Dmax |
5.0 |
nm |
| VolumePorod |
23 |
nm3 |
|
|