UniProt ID: P0ABT2 (None-None) DNA protection during starvation protein
|
|
|
|
| Sample: |
DNA protection during starvation protein dodecamer, 224 kDa Escherichia coli (strain … protein
|
| Buffer: |
10 mM Tris-HCl, 100 mM NaCl, 0.5 mM EDTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Oct 28
|
Polymorphic Protective Dps-DNA Co-Crystals by Cryo Electron Tomography and Small Angle X-Ray Scattering.
Biomolecules 10(1) (2019)
Kamyshinsky R, Chesnokov Y, Dadinova L, Mozhaev A, Orlov I, Petoukhov M, Orekhov A, Shtykova E, Vasiliev A
|
|
|
UniProt ID: A7ZJM7 (1-167) DNA protection during starvation protein
|
|
|

|
| Sample: |
DNA protection during starvation protein dodecamer, 225 kDa Escherichia coli protein
|
| Buffer: |
50 mM Tris-HCl, 50 mM NaCl, 0.5 mM EDTA, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Nov 26
|
Spatial Organization of Dps and DNA-Dps Complexes.
J Mol Biol :166930 (2021)
Dubrovin EV, Dadinova LA, Petoukhov MV, Yu Soshinskaya E, Mozhaev AA, Klinov DV, Schäffer TE, Shtykova EV, Batishchev OV
|
| RgGuinier |
3.9 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
314 |
nm3 |
|
|
UniProt ID: P0DTC1 (3860-3942) Replicase polyprotein 1a - nsp7
UniProt ID: P0DTC1 (3943-4140) Replicase polyprotein 1a - nsp8
|
|
|
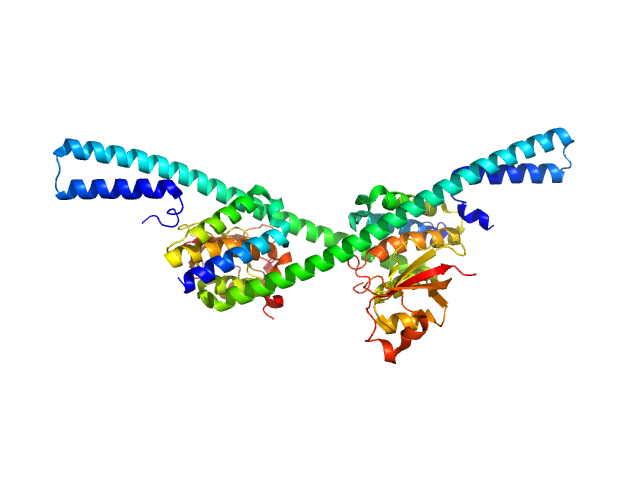
|
| Sample: |
Replicase polyprotein 1a - nsp7, 18 kDa Severe acute respiratory … protein
Replicase polyprotein 1a - nsp8, 44 kDa Severe acute respiratory … protein
|
| Buffer: |
50 mM Tris, pH 8.0, 100 mM NaCl, 4 mM DTT, 4 mM MgCl2, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Jun 19
|
Hallmarks of Alpha-
and Betacoronavirus
non-structural protein 7+8 complexes
Science Advances 7(10):eabf1004 (2021)
Krichel B, Bylapudi G, Schmidt C, Blanchet C, Schubert R, Brings L, Koehler M, Zenobi R, Svergun D, Lorenzen K, Madhugiri R, Ziebuhr J, Uetrecht C
|
| RgGuinier |
3.4 |
nm |
| Dmax |
13.5 |
nm |
| VolumePorod |
92 |
nm3 |
|
|
UniProt ID: O43464 (134-458) Serine protease HTRA2, mitochondrial
|
|
|
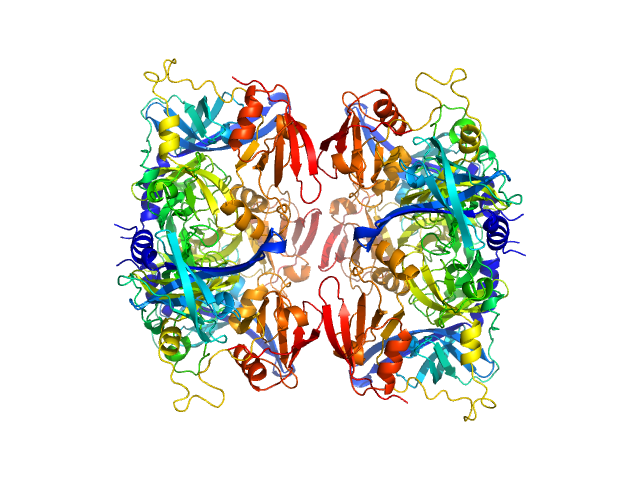
|
| Sample: |
Serine protease HTRA2, mitochondrial hexamer, 210 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES-NaOH (pH 7.4), 120 mM NaCl, 1 mM EDTA, and 2% glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2020 Jul 1
|
Oligomeric assembly regulating mitochondrial HtrA2 function as examined by methyl-TROSY NMR
Proceedings of the National Academy of Sciences 118(11):e2025022118 (2021)
Toyama Y, Harkness R, Lee T, Maynes J, Kay L
|
| RgGuinier |
4.2 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
250 |
nm3 |
|
|
UniProt ID: O43464 (134-458) Serine protease HTRA2, mitochondrial
|
|
|
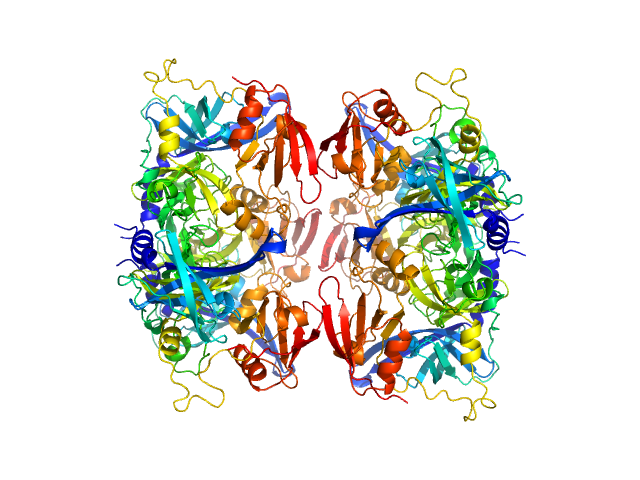
|
| Sample: |
Serine protease HTRA2, mitochondrial hexamer, 210 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES-NaOH (pH 7.4), 120 mM NaCl, 1 mM EDTA, and 2% glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2020 Jul 1
|
Oligomeric assembly regulating mitochondrial HtrA2 function as examined by methyl-TROSY NMR
Proceedings of the National Academy of Sciences 118(11):e2025022118 (2021)
Toyama Y, Harkness R, Lee T, Maynes J, Kay L
|
| RgGuinier |
3.8 |
nm |
| Dmax |
15.0 |
nm |
| VolumePorod |
213 |
nm3 |
|
|
UniProt ID: O43464 (134-458) Serine protease HTRA2, mitochondrial
|
|
|
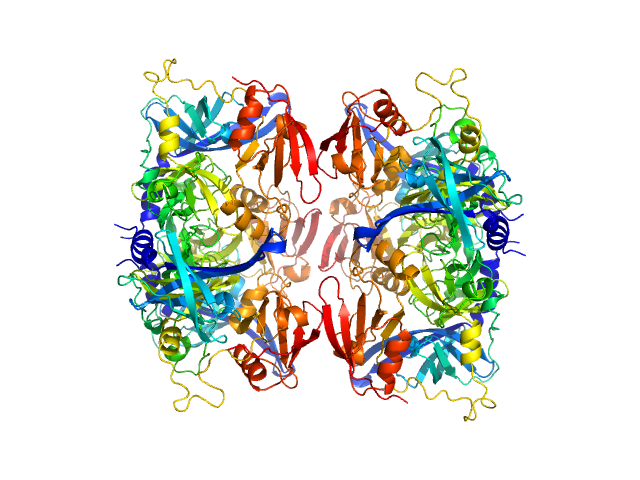
|
| Sample: |
Serine protease HTRA2, mitochondrial hexamer, 210 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES-NaOH (pH 7.4), 120 mM NaCl, 1 mM EDTA, and 2% glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2020 Jul 1
|
Oligomeric assembly regulating mitochondrial HtrA2 function as examined by methyl-TROSY NMR
Proceedings of the National Academy of Sciences 118(11):e2025022118 (2021)
Toyama Y, Harkness R, Lee T, Maynes J, Kay L
|
| RgGuinier |
3.6 |
nm |
| Dmax |
12.8 |
nm |
| VolumePorod |
203 |
nm3 |
|
|
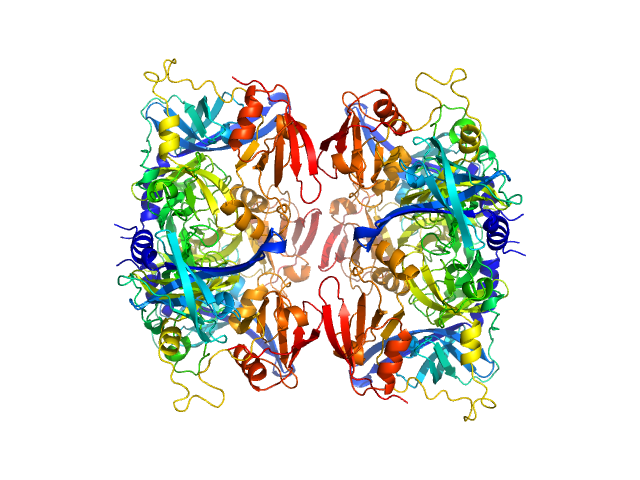
UniProt ID: O43464 (134-458) Serine protease HTRA2, mitochondrial
|
|
|
|
| Sample: |
Serine protease HTRA2, mitochondrial hexamer, 210 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES-NaOH (pH 7.4), 120 mM NaCl, 1 mM EDTA, and 2% glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2020 Jul 1
|
Oligomeric assembly regulating mitochondrial HtrA2 function as examined by methyl-TROSY NMR
Proceedings of the National Academy of Sciences 118(11):e2025022118 (2021)
Toyama Y, Harkness R, Lee T, Maynes J, Kay L
|
| RgGuinier |
3.4 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
168 |
nm3 |
|
|
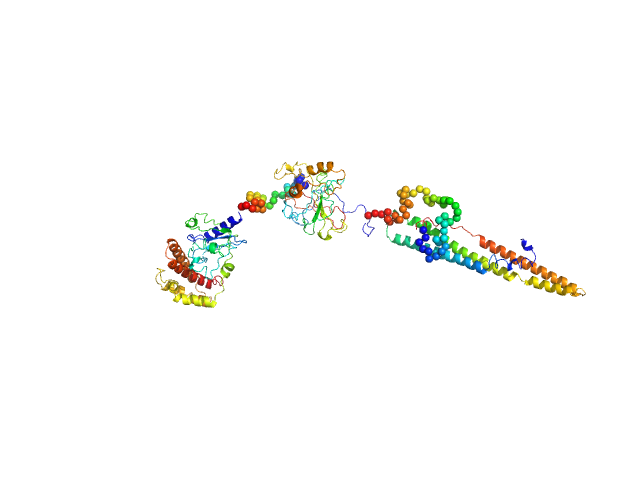
UniProt ID: P9WJC1 (484-729) ESX-1 secretion-associated protein EspK
|
|
|
|
| Sample: |
ESX-1 secretion-associated protein EspK monomer, 27 kDa Mycobacterium tuberculosis (strain … protein
|
| Buffer: |
20 mM Tris-HCl pH, 300 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Sep 20
|
Structural Analysis of the Partially Disordered Protein EspK from Mycobacterium Tuberculosis
Crystals 11(1):18 (2020)
Gijsbers A, Sánchez-Puig N, Gao Y, Peters P, Ravelli R, Siliqi D
|
| RgGuinier |
7.2 |
nm |
| Dmax |
3.3 |
nm |
| VolumePorod |
330 |
nm3 |
|
|
UniProt ID: P9WJC1 (517-729) ESX-1 secretion-associated protein EspK
|
|
|

|
| Sample: |
ESX-1 secretion-associated protein EspK monomer, 30 kDa Mycobacterium tuberculosis (strain … protein
|
| Buffer: |
20 mM Tris-HCl pH, 300 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Apr 12
|
Structural Analysis of the Partially Disordered Protein EspK from Mycobacterium Tuberculosis
Crystals 11(1):18 (2020)
Gijsbers A, Sánchez-Puig N, Gao Y, Peters P, Ravelli R, Siliqi D
|
| RgGuinier |
2.2 |
nm |
| Dmax |
8.4 |
nm |
| VolumePorod |
52 |
nm3 |
|
|
UniProt ID: Q6N022 (373-2769) Teneurin-4
|
|
|
|
| Sample: |
Teneurin-4 dimer, 537 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 7.8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Dec 16
|
Teneurin4 dimer structures reveal a calcium‐stabilized compact conformation supporting homomeric trans‐interactions
The EMBO Journal (2022)
Meijer D, Frias C, Beugelink J, Deurloo Y, Janssen B
|
| RgGuinier |
7.8 |
nm |
| Dmax |
32.0 |
nm |
| VolumePorod |
1280 |
nm3 |
|
|