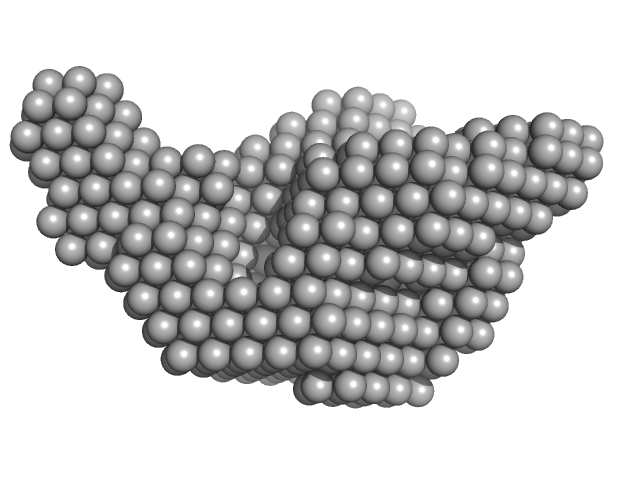
UniProt ID: F2X1X5 (1-103) Signal recognition particle 9
UniProt ID: F2X1X6 (1-104) Signal recognition particle 14
|
|
|
|
| Sample: |
Signal recognition particle 9 monomer, 12 kDa Plasmodium falciparum protein
Signal recognition particle 14 monomer, 12 kDa Plasmodium falciparum protein
|
| Buffer: |
20 mM HEPES pH 7.5, 150 mM NaCl, 10 mM MgCl2, 10 mM KCl, 1mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Feb 22
|
Structural analysis of the SRP Alu domain from Plasmodium falciparum reveals a non-canonical open conformation.
Commun Biol 4(1):600 (2021)
Soni K, Kempf G, Manalastas-Cantos K, Hendricks A, Flemming D, Guizetti J, Simon B, Frischknecht F, Svergun DI, Wild K, Sinning I
|
| RgGuinier |
2.1 |
nm |
| Dmax |
7.2 |
nm |
| VolumePorod |
47 |
nm3 |
|
|
UniProt ID: F2X1X5 (1-103) Signal recognition particle 9
UniProt ID: F2X1X6 (1-104) Signal recognition particle 14
UniProt ID: None (None-None) Full-length SRP Alu RNA
|
|
|

|
| Sample: |
Signal recognition particle 9 monomer, 12 kDa Plasmodium falciparum protein
Signal recognition particle 14 monomer, 12 kDa Plasmodium falciparum protein
Full-length SRP Alu RNA monomer, 38 kDa Plasmodium falciparum RNA
|
| Buffer: |
20 mM HEPES pH 7.5, 150 mM NaCl, 10 mM MgCl2, 10 mM KCl, 1mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Jun 22
|
Structural analysis of the SRP Alu domain from Plasmodium falciparum reveals a non-canonical open conformation.
Commun Biol 4(1):600 (2021)
Soni K, Kempf G, Manalastas-Cantos K, Hendricks A, Flemming D, Guizetti J, Simon B, Frischknecht F, Svergun DI, Wild K, Sinning I
|
| RgGuinier |
3.5 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
120 |
nm3 |
|
|
UniProt ID: F2X1X5 (1-103) Signal recognition particle 9
UniProt ID: F2X1X6 (1-104) Signal recognition particle 14
UniProt ID: None (None-None) SRP Alu RNA 5' domain
|
|
|

|
| Sample: |
Signal recognition particle 9 monomer, 12 kDa Plasmodium falciparum protein
Signal recognition particle 14 monomer, 12 kDa Plasmodium falciparum protein
SRP Alu RNA 5' domain monomer, 24 kDa Plasmodium falciparum RNA
|
| Buffer: |
20 mM HEPES pH 7.5, 150 mM NaCl, 10 mM MgCl2, 10 mM KCl, 1mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Jun 22
|
Structural analysis of the SRP Alu domain from Plasmodium falciparum reveals a non-canonical open conformation.
Commun Biol 4(1):600 (2021)
Soni K, Kempf G, Manalastas-Cantos K, Hendricks A, Flemming D, Guizetti J, Simon B, Frischknecht F, Svergun DI, Wild K, Sinning I
|
| RgGuinier |
3.2 |
nm |
| Dmax |
11.9 |
nm |
| VolumePorod |
77 |
nm3 |
|
|
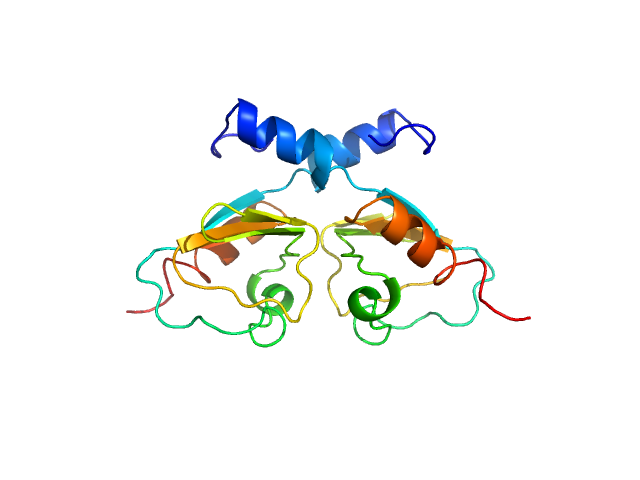
UniProt ID: Q8E533 (134-213) Transcriptional repressor BusR RCK_C domain
|
|
|
|
| Sample: |
Transcriptional repressor BusR RCK_C domain dimer, 22 kDa Streptococcus agalactiae serotype … protein
|
| Buffer: |
100 mM NaCl, 30mM Hepes, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Jul 2
|
BusR senses bipartite DNA binding motifs by a unique molecular ruler architecture.
Nucleic Acids Res (2021)
Bandera AM, Bartho J, Lammens K, Drexler DJ, Kleinschwärzer J, Hopfner KP, Witte G
|
| RgGuinier |
1.9 |
nm |
| Dmax |
6.4 |
nm |
| VolumePorod |
44 |
nm3 |
|
|
UniProt ID: Q8E533 (1-213) Transcriptional repressor BusR
|
|
|
|
| Sample: |
Transcriptional repressor BusR tetramer, 95 kDa Streptococcus agalactiae protein
|
| Buffer: |
20mM HEPES, pH6.5, 100mM NaCl, 3% glycerol (v/v), pH: 6.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Jul 2
|
BusR senses bipartite DNA binding motifs by a unique molecular ruler architecture.
Nucleic Acids Res (2021)
Bandera AM, Bartho J, Lammens K, Drexler DJ, Kleinschwärzer J, Hopfner KP, Witte G
|
| RgGuinier |
4.4 |
nm |
| Dmax |
13.9 |
nm |
| VolumePorod |
168 |
nm3 |
|
|
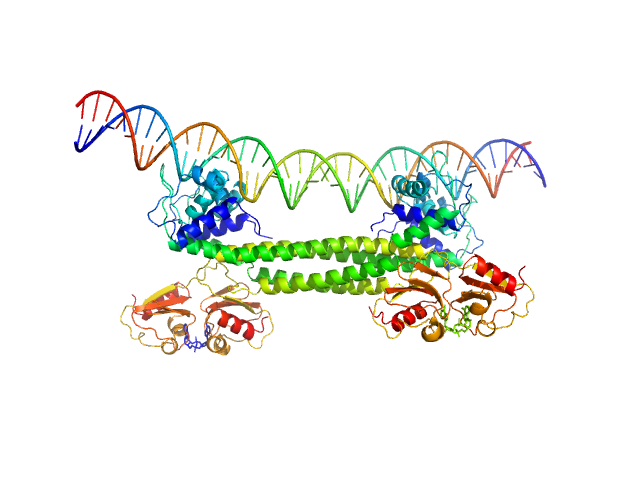
UniProt ID: Q8E533 (1-213) Transcriptional repressor BusR
UniProt ID: None (None-None) BusR Recognition sequence
|
|
|
|
| Sample: |
Transcriptional repressor BusR tetramer, 95 kDa Streptococcus agalactiae protein
BusR Recognition sequence monomer, 28 kDa synthetic construct DNA
|
| Buffer: |
20mM HEPES, pH6.5, 100mM NaCl, 3% glycerol (v/v), pH: 6.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Jul 2
|
BusR senses bipartite DNA binding motifs by a unique molecular ruler architecture.
Nucleic Acids Res (2021)
Bandera AM, Bartho J, Lammens K, Drexler DJ, Kleinschwärzer J, Hopfner KP, Witte G
|
| RgGuinier |
4.3 |
nm |
| Dmax |
14.2 |
nm |
| VolumePorod |
210 |
nm3 |
|
|
UniProt ID: P27918 (28-469) Properdin (dimer)
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDKA4_fit1_model1.png)
|
| Sample: |
Properdin (dimer) dimer, 110 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Nov 14
|
Properdin oligomers adopt rigid extended conformations supporting function.
Elife 10 (2021)
Pedersen DV, Pedersen MN, Mazarakis SM, Wang Y, Lindorff-Larsen K, Arleth L, Andersen GR
|
| RgGuinier |
8.1 |
nm |
| Dmax |
24.0 |
nm |
|
|
UniProt ID: P27918 (28-469) Properdin (trimer)
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDKB4_fit1_model1.png)
|
| Sample: |
Properdin (trimer) trimer, 165 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Nov 14
|
Properdin oligomers adopt rigid extended conformations supporting function.
Elife 10 (2021)
Pedersen DV, Pedersen MN, Mazarakis SM, Wang Y, Lindorff-Larsen K, Arleth L, Andersen GR
|
| RgGuinier |
10.2 |
nm |
| Dmax |
27.0 |
nm |
|
|
UniProt ID: P27918 (28-469) Properdin (tetramer)
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDKC4_fit1_model1.png)
|
| Sample: |
Properdin (tetramer) tetramer, 220 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Nov 14
|
Properdin oligomers adopt rigid extended conformations supporting function.
Elife 10 (2021)
Pedersen DV, Pedersen MN, Mazarakis SM, Wang Y, Lindorff-Larsen K, Arleth L, Andersen GR
|
| RgGuinier |
13.1 |
nm |
| Dmax |
36.0 |
nm |
|
|
UniProt ID: P0ABT2 (None-None) DNA protection during starvation protein
|
|
|
|
| Sample: |
DNA protection during starvation protein dodecamer, 224 kDa Escherichia coli (strain … protein
|
| Buffer: |
50 mM Tris-HCl, 50 mM NaCl, 0.5 mM EDTA, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Nov 27
|
Protective Dps-DNA co-crystallization in stressed cells: an in vitro structural study by small-angle X-ray scattering and cryo-electron tomography.
FEBS Lett 593(12):1360-1371 (2019)
Dadinova LA, Chesnokov YM, Kamyshinsky RA, Orlov IA, Petoukhov MV, Mozhaev AA, Soshinskaya EY, Lazarev VN, Manuvera VA, Orekhov AS, Vasiliev AL, Shtykova EV
|
|
|