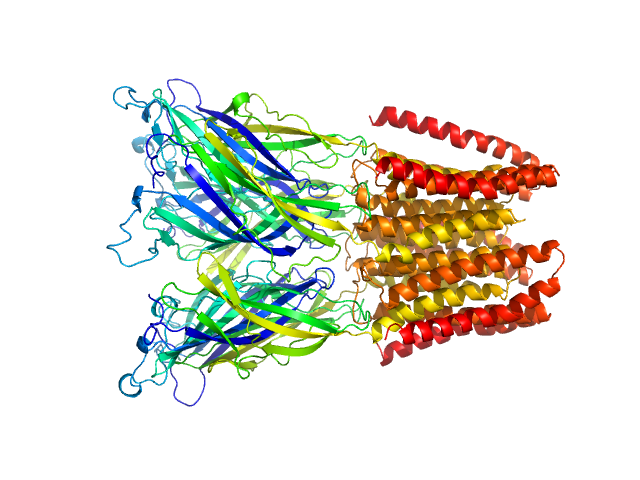
UniProt ID: Q7NDN8 (44-359) Proton-gated ion channel
|
|
|
|
| Sample: |
Proton-gated ion channel pentamer, 183 kDa Gloeobacter violaceus (strain … protein
|
| Buffer: |
D2O, 20 mM Tris, 150 mM NaCl, 0.5 mM matched-out deuterated DDM,, pH: 7.5 |
| Experiment: |
SANS
data collected at D22, Institut Laue-Langevin (ILL) on 2020 Aug 22
|
Probing solution structure of the pentameric ligand-gated ion channel GLIC by small-angle neutron scattering
Proceedings of the National Academy of Sciences 118(37):e2108006118 (2021)
Lycksell M, Rovšnik U, Bergh C, Johansen N, Martel A, Porcar L, Arleth L, Howard R, Lindahl E
|
| RgGuinier |
3.8 |
nm |
| Dmax |
13.5 |
nm |
| VolumePorod |
274 |
nm3 |
|
|
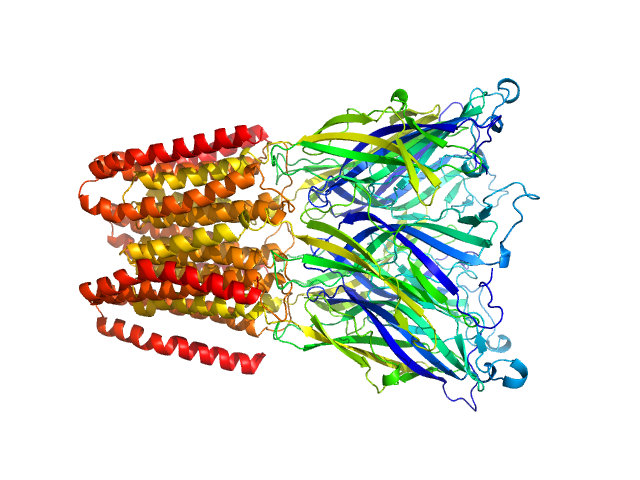
UniProt ID: Q7NDN8 (44-359) Proton-gated ion channel
|
|
|
|
| Sample: |
Proton-gated ion channel pentamer, 183 kDa Gloeobacter violaceus (strain … protein
|
| Buffer: |
D2O, 20 mM citrate, 150 mM NaCl, 0.5 mM match-out deuterated DDM, pH: 3 |
| Experiment: |
SANS
data collected at D22, Institut Laue-Langevin (ILL) on 2020 Aug 22
|
Probing solution structure of the pentameric ligand-gated ion channel GLIC by small-angle neutron scattering
Proceedings of the National Academy of Sciences 118(37):e2108006118 (2021)
Lycksell M, Rovšnik U, Bergh C, Johansen N, Martel A, Porcar L, Arleth L, Howard R, Lindahl E
|
| RgGuinier |
3.8 |
nm |
| Dmax |
12.7 |
nm |
| VolumePorod |
279 |
nm3 |
|
|
UniProt ID: Q7NDN8 (44-359) Proton-gated ion channel
|
|
|
|
| Sample: |
Proton-gated ion channel pentamer, 183 kDa Gloeobacter violaceus (strain … protein
|
| Buffer: |
D2O, 20 mM Tris, 150 mM NaCl, 0.5 mM matched-out deuterated DDM,, pH: 7.5 |
| Experiment: |
SANS
data collected at D22, Institut Laue-Langevin (ILL) on 2019 Jun 20
|
Probing solution structure of the pentameric ligand-gated ion channel GLIC by small-angle neutron scattering
Proceedings of the National Academy of Sciences 118(37):e2108006118 (2021)
Lycksell M, Rovšnik U, Bergh C, Johansen N, Martel A, Porcar L, Arleth L, Howard R, Lindahl E
|
| RgGuinier |
3.8 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
235 |
nm3 |
|
|
UniProt ID: Q7NDN8 (44-359) Proton-gated ion channel
|
|
|
|
| Sample: |
Proton-gated ion channel pentamer, 183 kDa Gloeobacter violaceus (strain … protein
|
| Buffer: |
D2O, 20 mM Tris, 150 mM NaCl, 0.5 mM matched-out deuterated DDM,, pH: 7.5 |
| Experiment: |
SANS
data collected at D22, Institut Laue-Langevin (ILL) on 2019 Jun 21
|
Probing solution structure of the pentameric ligand-gated ion channel GLIC by small-angle neutron scattering
Proceedings of the National Academy of Sciences 118(37):e2108006118 (2021)
Lycksell M, Rovšnik U, Bergh C, Johansen N, Martel A, Porcar L, Arleth L, Howard R, Lindahl E
|
| RgGuinier |
4.0 |
nm |
| Dmax |
17.7 |
nm |
| VolumePorod |
225 |
nm3 |
|
|
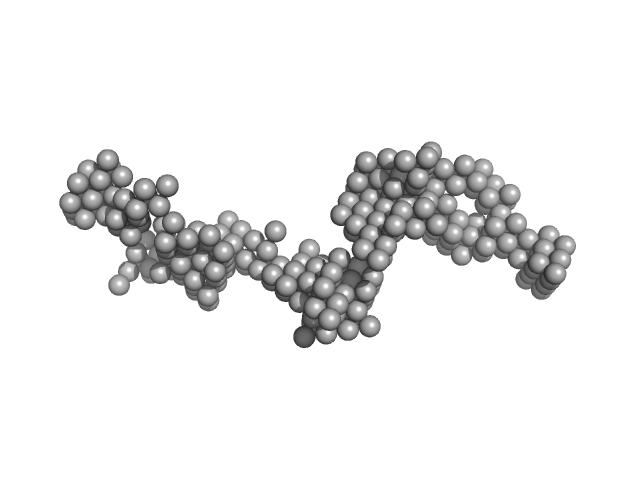
UniProt ID: Q70AB8 (28-266) Protein A
|
|
|
|
| Sample: |
Protein A monomer, 28 kDa Staphylococcus aureus protein
|
| Buffer: |
10 mM HEPES, pH 7.2, 3 mM EDTA, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2005 May 12
|
A structural basis for Staphylococcal complement subversion: X-ray structure of the complement-binding domain of Staphylococcus aureus protein Sbi in complex with ligand C3d
Molecular Immunology 48(4):452-462 (2011)
Clark E, Crennell S, Upadhyay A, Zozulya A, Mackay J, Svergun D, Bagby S, van den Elsen J
|
| RgGuinier |
4.6 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
81 |
nm3 |
|
|
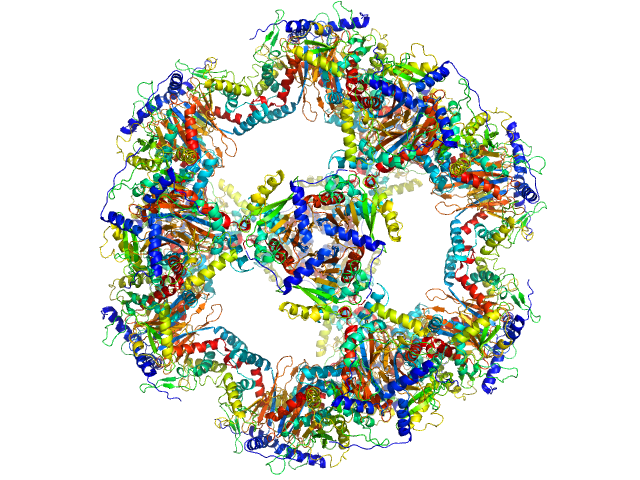
UniProt ID: P03120 (177-400) Regulatory protein E2
|
|
|
|
| Sample: |
Regulatory protein E2, 1037 kDa Human papillomavirus type … protein
|
| Buffer: |
50 mM Tris ⁄ HCl, pH 8.8, 100 mM NaCl, pH: 8.8 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2009 Jul 13
|
The catalytic core of an archaeal 2-oxoacid dehydrogenase multienzyme complex is a 42-mer protein assembly
FEBS Journal 279(5):713-723 (2012)
Marrott N, Marshall J, Svergun D, Crennell S, Hough D, Danson M, van den Elsen J
|
| RgGuinier |
8.8 |
nm |
| Dmax |
22.0 |
nm |
| VolumePorod |
2473 |
nm3 |
|
|
UniProt ID: P03120 (182-400) Regulatory protein E2
|
|
|
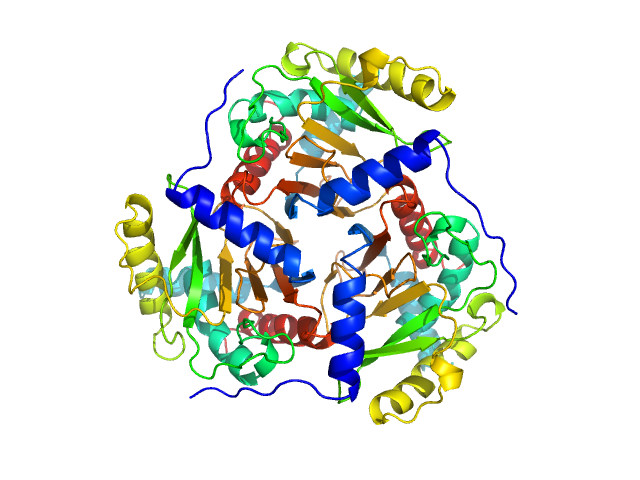
|
| Sample: |
Regulatory protein E2 trimer, 73 kDa Human papillomavirus type … protein
|
| Buffer: |
50 mM Tris ⁄ HCl, pH 8.8, 100 mM NaCl, pH: 8.8 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 May 27
|
Why are the 2-oxoacid dehydrogenase complexes so large? Generation of an active trimeric complex
Biochemical Journal 463(3):405-412 (2014)
Marrott N, Marshall J, Svergun D, Crennell S, Hough D, van den Elsen J, Danson M
|
| RgGuinier |
3.1 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
149 |
nm3 |
|
|
UniProt ID: Q9JLU4 (1-676) SH3 and multiple ankyrin repeat domains protein 3
|
|
|
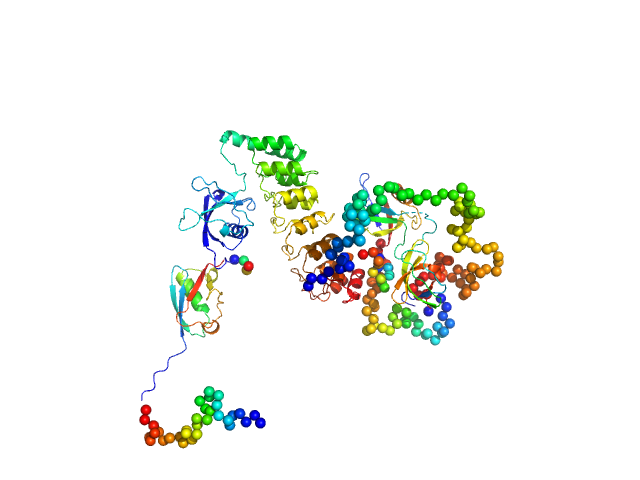
|
| Sample: |
SH3 and multiple ankyrin repeat domains protein 3 monomer, 88 kDa Rattus norvegicus protein
|
| Buffer: |
100mM NaH2PO4, 100mM NaCl, 0.5mM DTT,, pH: 6.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Jun 3
|
Autism associated SHANK3 missense point mutations impact conformational fluctuations and protein turnover at synapses.
Elife 10 (2021)
Bucher M, Niebling S, Han Y, Molodenskiy D, Nia FH, Kreienkamp HJ, Svergun D, Kim E, Kostyukova AS, Kreutz MR, Mikhaylova M
|
| RgGuinier |
4.1 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
170 |
nm3 |
|
|
UniProt ID: Q9JLU4 (1-676) SH3 and multiple ankyrin repeat domains protein 3
|
|
|
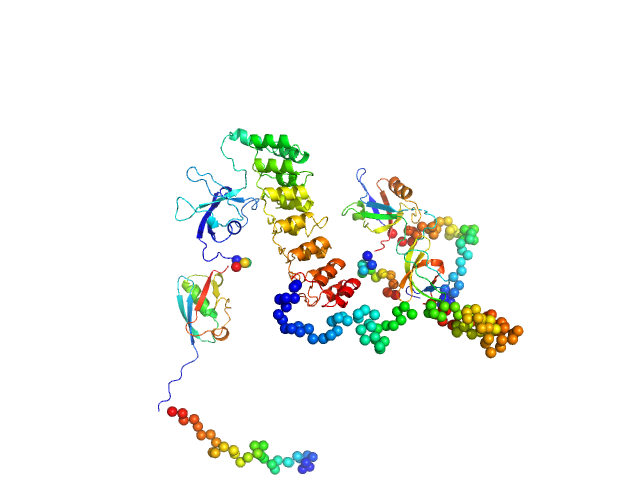
|
| Sample: |
SH3 and multiple ankyrin repeat domains protein 3 monomer, 88 kDa Rattus norvegicus protein
|
| Buffer: |
100mM NaH2PO4, 100mM NaCl, 0.5mM DTT,, pH: 6.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Jun 3
|
Autism associated SHANK3 missense point mutations impact conformational fluctuations and protein turnover at synapses.
Elife 10 (2021)
Bucher M, Niebling S, Han Y, Molodenskiy D, Nia FH, Kreienkamp HJ, Svergun D, Kim E, Kostyukova AS, Kreutz MR, Mikhaylova M
|
| RgGuinier |
4.1 |
nm |
| Dmax |
14.8 |
nm |
| VolumePorod |
224 |
nm3 |
|
|
UniProt ID: Q9JLU4 (1-676) SH3 and multiple ankyrin repeat domains protein 3
|
|
|
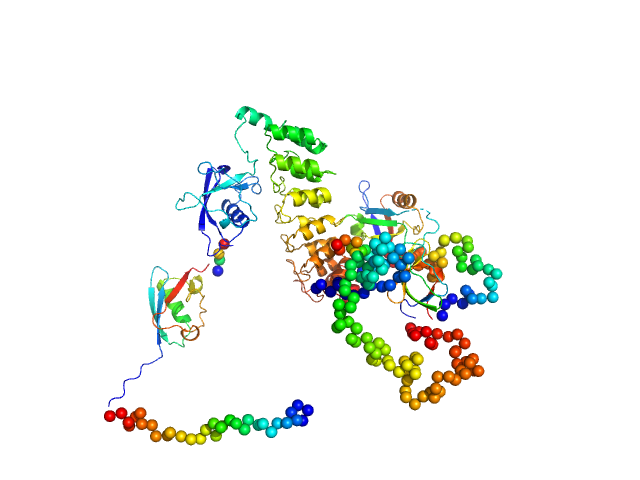
|
| Sample: |
SH3 and multiple ankyrin repeat domains protein 3 monomer, 87 kDa Rattus norvegicus protein
|
| Buffer: |
100mM NaH2PO4, 100mM NaCl, 0.5mM DTT,, pH: 6.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Jun 3
|
Autism associated SHANK3 missense point mutations impact conformational fluctuations and protein turnover at synapses.
Elife 10 (2021)
Bucher M, Niebling S, Han Y, Molodenskiy D, Nia FH, Kreienkamp HJ, Svergun D, Kim E, Kostyukova AS, Kreutz MR, Mikhaylova M
|
| RgGuinier |
4.1 |
nm |
| Dmax |
13.8 |
nm |
| VolumePorod |
175 |
nm3 |
|
|