UniProt ID: P10636 (504-758) Microtubule-associated protein tau, Tau35 fragment
|
|
|
|
| Sample: |
Microtubule-associated protein tau, Tau35 fragment monomer, 27 kDa Homo sapiens protein
|
| Buffer: |
phosphate buffered saline, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Oct 8
|
The Disease Associated Tau35 Fragment has an Increased Propensity to Aggregate Compared to Full-Length Tau
Frontiers in Molecular Biosciences 8 (2021)
Lyu C, Da Vela S, Al-Hilaly Y, Marshall K, Thorogate R, Svergun D, Serpell L, Pastore A, Hanger D
|
| RgGuinier |
4.6 |
nm |
| Dmax |
16.2 |
nm |
| VolumePorod |
93 |
nm3 |
|
|
UniProt ID: P10636 (None-None) Microtubule-associated protein tau, 2N3R isoform
|
|
|
|
| Sample: |
Microtubule-associated protein tau, 2N3R isoform monomer, 43 kDa Homo sapiens protein
|
| Buffer: |
phosphate buffered saline, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Oct 8
|
The Disease Associated Tau35 Fragment has an Increased Propensity to Aggregate Compared to Full-Length Tau
Frontiers in Molecular Biosciences 8 (2021)
Lyu C, Da Vela S, Al-Hilaly Y, Marshall K, Thorogate R, Svergun D, Serpell L, Pastore A, Hanger D
|
| RgGuinier |
6.3 |
nm |
| Dmax |
19.8 |
nm |
| VolumePorod |
211 |
nm3 |
|
|
UniProt ID: P10636 (None-None) Microtubule-associated protein tau, 2N4R isoform
|
|
|
|
| Sample: |
Microtubule-associated protein tau, 2N4R isoform monomer, 46 kDa Homo sapiens protein
|
| Buffer: |
phosphate buffered saline, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Oct 8
|
The Disease Associated Tau35 Fragment has an Increased Propensity to Aggregate Compared to Full-Length Tau
Frontiers in Molecular Biosciences 8 (2021)
Lyu C, Da Vela S, Al-Hilaly Y, Marshall K, Thorogate R, Svergun D, Serpell L, Pastore A, Hanger D
|
| RgGuinier |
6.7 |
nm |
| Dmax |
23.5 |
nm |
| VolumePorod |
254 |
nm3 |
|
|
UniProt ID: P00450 (1-1065) Ceruloplasmin
UniProt ID: P02788 (1-710) Lactotransferrin
UniProt ID: P05164 (167-745) Myeloperoxidase
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDLV4_fit1_model1.png)
|
| Sample: |
Ceruloplasmin dimer, 244 kDa Homo sapiens protein
Lactotransferrin dimer, 156 kDa Homo sapiens protein
Myeloperoxidase dimer, 132 kDa Homo sapiens protein
|
| Buffer: |
0.1 M sodium-acetate buffer, pH: 5.6 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2007 Nov 24
|
Ceruloplasmin: Macromolecular Assemblies with Iron-Containing Acute Phase Proteins
PLoS ONE 8(7):e67145 (2013)
Samygina V, Sokolov A, Bourenkov G, Petoukhov M, Pulina M, Zakharova E, Vasilyev V, Bartunik H, Svergun D, Tuma R
|
| RgGuinier |
6.9 |
nm |
| Dmax |
22.6 |
nm |
| VolumePorod |
726 |
nm3 |
|
|
UniProt ID: P26447 (1-101) Protein S100-A4
|
|
|
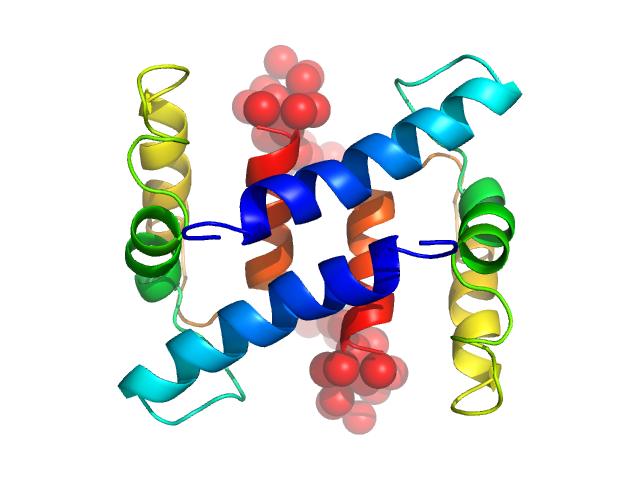
|
| Sample: |
Protein S100-A4 dimer, 23 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 20 mM NaCl, 0.1 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2013 Mar 2
|
The C-Terminal Random Coil Region Tunes the Ca2+-Binding Affinity of S100A4 through Conformational Activation
PLoS ONE 9(5):e97654 (2014)
Duelli A, Kiss B, Lundholm I, Bodor A, Petoukhov M, Svergun D, Nyitray L, Katona G, Csernoch L
|
| RgGuinier |
2.1 |
nm |
| Dmax |
7.0 |
nm |
| VolumePorod |
48 |
nm3 |
|
|
UniProt ID: Q46877 (1-479) Anaerobic nitric oxide reductase flavorubredoxin
|
|
|
|
| Sample: |
Anaerobic nitric oxide reductase flavorubredoxin tetramer, 217 kDa Escherichia coli (strain … protein
|
| Buffer: |
50 mM Tris-HCl, 18% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2006 Nov 19
|
Quaternary Structure of Flavorubredoxin as Revealed by Synchrotron Radiation Small-Angle X-Ray Scattering
Structure 16(9):1428-1436 (2008)
Petoukhov M, Vicente J, Crowley P, Carrondo M, Teixeira M, Svergun D
|
| RgGuinier |
4.1 |
nm |
| Dmax |
14.2 |
nm |
| VolumePorod |
297 |
nm3 |
|
|
UniProt ID: Q09808 (1-117) Survival motor neuron-like protein 1
UniProt ID: P0CU08 (1-235) Survival of motor neuron protein-interacting protein yip11
UniProt ID: None (249-281) Gcn4p
|
|
|
|
| Sample: |
Survival motor neuron-like protein 1 dimer, 26 kDa Schizosaccharomyces pombe (strain … protein
Survival of motor neuron protein-interacting protein yip11 dimer, 54 kDa Schizosaccharomyces pombe (strain … protein
Gcn4p dimer, 8 kDa Saccharomyces cerevisiae (strain … protein
|
| Buffer: |
20 mM Na/KPO4 pH 7.0, 150 mM NaCl, 1 mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at X21, National Synchrotron Light Source (NSLS) on 2011 Oct 21
|
Assembly of higher-order SMN oligomers is essential for metazoan viability and requires an exposed structural motif present in the YG zipper dimer.
Nucleic Acids Res 49(13):7644-7664 (2021)
Gupta K, Wen Y, Ninan NS, Raimer AC, Sharp R, Spring AM, Sarachan KL, Johnson MC, Van Duyne GD, Matera AG
|
| RgGuinier |
6.6 |
nm |
| Dmax |
23.3 |
nm |
| VolumePorod |
320 |
nm3 |
|
|
UniProt ID: Q09808 (1-117) Survival motor neuron-like protein 1
UniProt ID: P0CU08 (1-235) Survival of motor neuron protein-interacting protein yip11
UniProt ID: None (249-281) Gcn4p
|
|
|
|
| Sample: |
Survival motor neuron-like protein 1 dimer, 26 kDa Schizosaccharomyces pombe (strain … protein
Survival of motor neuron protein-interacting protein yip11 dimer, 54 kDa Schizosaccharomyces pombe (strain … protein
Gcn4p dimer, 8 kDa Saccharomyces cerevisiae (strain … protein
|
| Buffer: |
20 mM Na/KPO4 pH 7.0, 150 mM NaCl, 1 mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at X21, National Synchrotron Light Source (NSLS) on 2011 Oct 21
|
Assembly of higher-order SMN oligomers is essential for metazoan viability and requires an exposed structural motif present in the YG zipper dimer.
Nucleic Acids Res 49(13):7644-7664 (2021)
Gupta K, Wen Y, Ninan NS, Raimer AC, Sharp R, Spring AM, Sarachan KL, Johnson MC, Van Duyne GD, Matera AG
|
| RgGuinier |
6.6 |
nm |
| Dmax |
22.9 |
nm |
| VolumePorod |
325 |
nm3 |
|
|
UniProt ID: Q09808 (1-117) Survival motor neuron-like protein 1
UniProt ID: P0CU08 (1-235) Survival of motor neuron protein-interacting protein yip11
UniProt ID: None (249-281) Gcn4p
|
|
|
|
| Sample: |
Survival motor neuron-like protein 1 dimer, 26 kDa Schizosaccharomyces pombe (strain … protein
Survival of motor neuron protein-interacting protein yip11 dimer, 54 kDa Schizosaccharomyces pombe (strain … protein
Gcn4p dimer, 8 kDa Saccharomyces cerevisiae (strain … protein
|
| Buffer: |
20 mM Na/KPO4 pH 7.0, 150 mM NaCl, 1 mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at X21, National Synchrotron Light Source (NSLS) on 2011 Oct 21
|
Assembly of higher-order SMN oligomers is essential for metazoan viability and requires an exposed structural motif present in the YG zipper dimer.
Nucleic Acids Res 49(13):7644-7664 (2021)
Gupta K, Wen Y, Ninan NS, Raimer AC, Sharp R, Spring AM, Sarachan KL, Johnson MC, Van Duyne GD, Matera AG
|
| RgGuinier |
6.4 |
nm |
| Dmax |
21.9 |
nm |
| VolumePorod |
315 |
nm3 |
|
|
UniProt ID: Q09808 (1-117) Survival motor neuron-like protein 1
UniProt ID: P0CU08 (1-235) Survival of motor neuron protein-interacting protein yip11
UniProt ID: None (249-281) Gcn4p
|
|
|
|
| Sample: |
Survival motor neuron-like protein 1 dimer, 26 kDa Schizosaccharomyces pombe (strain … protein
Survival of motor neuron protein-interacting protein yip11 dimer, 54 kDa Schizosaccharomyces pombe (strain … protein
Gcn4p dimer, 8 kDa Saccharomyces cerevisiae (strain … protein
|
| Buffer: |
20 mM Na/KPO4 pH 7.0, 150 mM NaCl, 1 mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at X21, National Synchrotron Light Source (NSLS) on 2011 Oct 21
|
Assembly of higher-order SMN oligomers is essential for metazoan viability and requires an exposed structural motif present in the YG zipper dimer.
Nucleic Acids Res 49(13):7644-7664 (2021)
Gupta K, Wen Y, Ninan NS, Raimer AC, Sharp R, Spring AM, Sarachan KL, Johnson MC, Van Duyne GD, Matera AG
|
| RgGuinier |
6.3 |
nm |
| Dmax |
19.9 |
nm |
| VolumePorod |
315 |
nm3 |
|
|