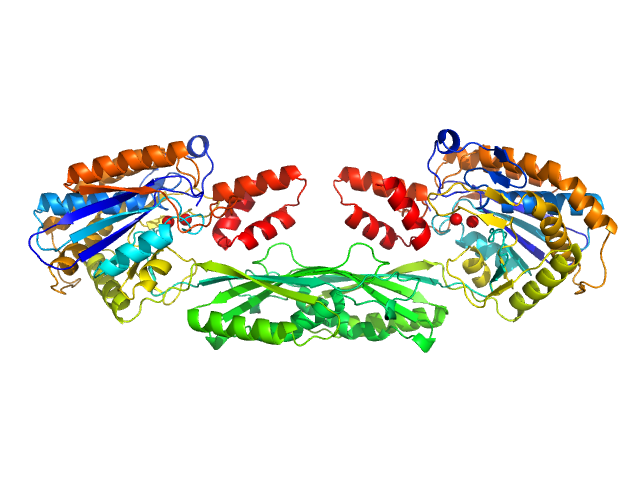
UniProt ID: P06621 (89-415) Pro-Carboxypeptidase G2 (circular permutant CP-N89) K177A Design 3
|
|
|
|
| Sample: |
Pro-Carboxypeptidase G2 (circular permutant CP-N89) K177A Design 3 dimer, 100 kDa Pseudomonas sp. (strain … protein
|
| Buffer: |
50 mM Tris, 100 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 May 8
|
Massively parallel, computationally guided design of a proenzyme.
Proc Natl Acad Sci U S A 119(15):e2116097119 (2022)
Yachnin BJ, Azouz LR, White RE 3rd, Minetti CASA, Remeta DP, Tan VM, Drake JM, Khare SD
|
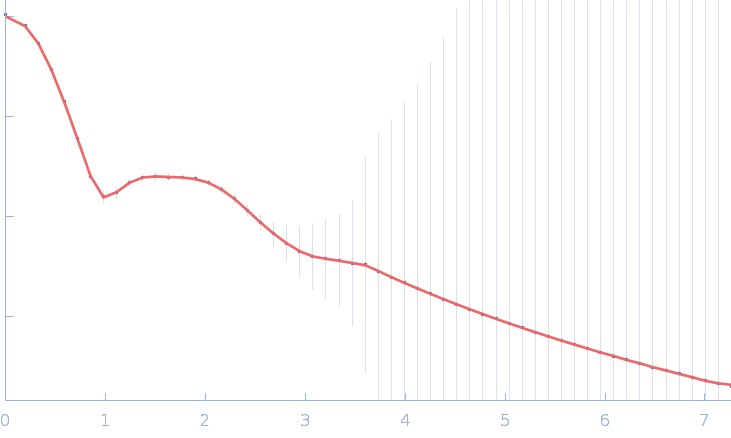
| RgGuinier |
3.7 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
127 |
nm3 |
|
|
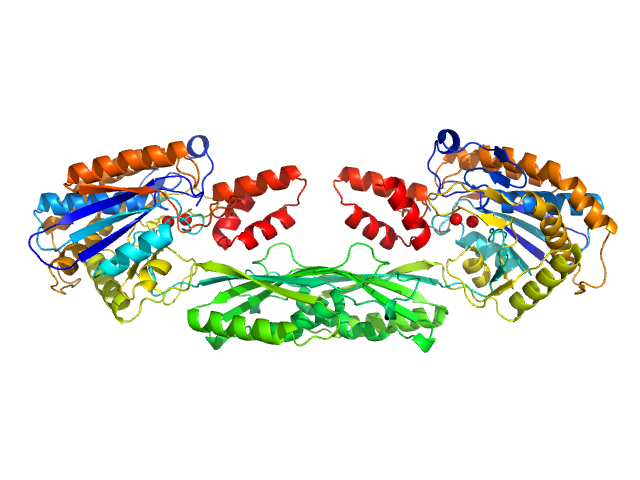
UniProt ID: None (None-None) Methotrexate
UniProt ID: P06621 (89-415) Pro-Carboxypeptidase G2 (circular permutant CP-N89) K177A Design 3
|
|
|
|
| Sample: |
Methotrexate dimer, 1 kDa
Pro-Carboxypeptidase G2 (circular permutant CP-N89) K177A Design 3 dimer, 100 kDa Pseudomonas sp. (strain … protein
|
| Buffer: |
50 mM Tris, 100 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 May 8
|
Massively parallel, computationally guided design of a proenzyme.
Proc Natl Acad Sci U S A 119(15):e2116097119 (2022)
Yachnin BJ, Azouz LR, White RE 3rd, Minetti CASA, Remeta DP, Tan VM, Drake JM, Khare SD
|
| RgGuinier |
3.5 |
nm |
| Dmax |
12.3 |
nm |
| VolumePorod |
125 |
nm3 |
|
|
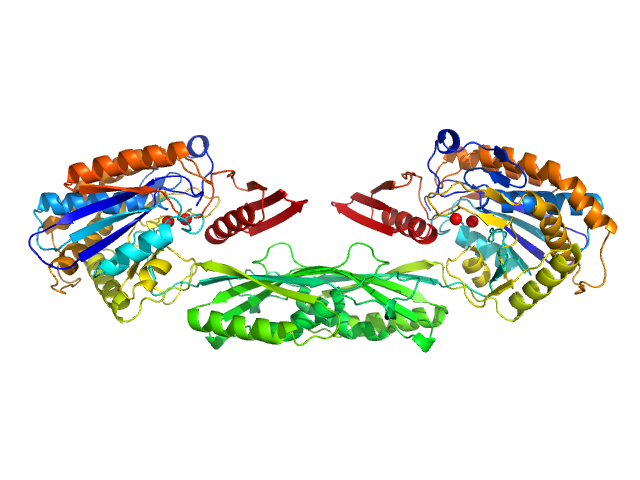
UniProt ID: P06621 (89-415) Pro-Carboxypeptidase G2 (circular permutant CP-N89) K177A Design 1 Disulfide Variant
|
|
|
|
| Sample: |
Pro-Carboxypeptidase G2 (circular permutant CP-N89) K177A Design 1 Disulfide Variant dimer, 96 kDa Pseudomonas sp. (strain … protein
|
| Buffer: |
50 mM Tris, 100 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 Dec 6
|
Massively parallel, computationally guided design of a proenzyme.
Proc Natl Acad Sci U S A 119(15):e2116097119 (2022)
Yachnin BJ, Azouz LR, White RE 3rd, Minetti CASA, Remeta DP, Tan VM, Drake JM, Khare SD
|
| RgGuinier |
3.6 |
nm |
| Dmax |
12.6 |
nm |
| VolumePorod |
120 |
nm3 |
|
|
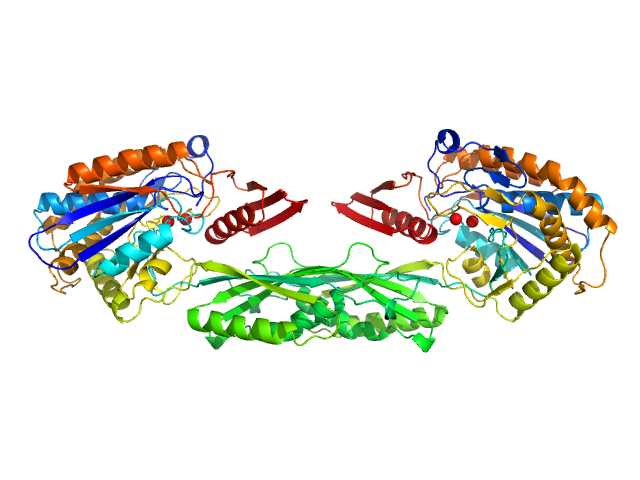
UniProt ID: None (None-None) Methotrexate
UniProt ID: P06621 (89-415) Pro-Carboxypeptidase G2 (circular permutant CP-N89) K177A Design 1 Disulfide Variant
|
|
|
|
| Sample: |
Methotrexate dimer, 1 kDa
Pro-Carboxypeptidase G2 (circular permutant CP-N89) K177A Design 1 Disulfide Variant dimer, 96 kDa Pseudomonas sp. (strain … protein
|
| Buffer: |
50 mM Tris, 100 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 Dec 6
|
Massively parallel, computationally guided design of a proenzyme.
Proc Natl Acad Sci U S A 119(15):e2116097119 (2022)
Yachnin BJ, Azouz LR, White RE 3rd, Minetti CASA, Remeta DP, Tan VM, Drake JM, Khare SD
|
| RgGuinier |
3.5 |
nm |
| Dmax |
12.3 |
nm |
| VolumePorod |
125 |
nm3 |
|
|
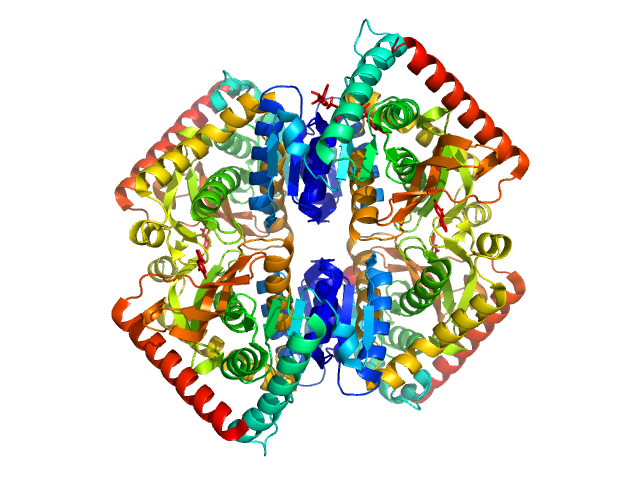
UniProt ID: Q5NH85 (24-393) Francisella tularensis outer membrane protein A
|
|
|
|
| Sample: |
Francisella tularensis outer membrane protein A dimer, 80 kDa Francisella tularensis subsp. … protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 0.05% B-DDM, pH: 7.5 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2019 Mar 20
|
Structural and biophysical properties of FopA, a major outer membrane protein of Francisella tularensis.
PLoS One 17(8):e0267370 (2022)
Nagaratnam N, Martin-Garcia JM, Yang JH, Goode MR, Ketawala G, Craciunescu FM, Zook JD, Sonowal M, Williams D, Grant TD, Fromme R, Hansen DT, Fromme P
|
| RgGuinier |
4.4 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
330 |
nm3 |
|
|
UniProt ID: Q6VVP7 (1-313) L-lactate dehydrogenase
|
|
|
|
| Sample: |
L-lactate dehydrogenase tetramer, 141 kDa Plasmodium falciparum protein
|
| Buffer: |
100 mM Na-phosphate buffer, 400 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at Xenocs Xeuss 2.0 with MetalJet, Department of Macromolecular Physics, Faculty of Physics, Adam Mickiewicz University on 2019 Jul 3
|
A fragment-based approach identifies an allosteric pocket that impacts malate dehydrogenase activity
Communications Biology 4(1) (2021)
Reyes Romero A, Lunev S, Popowicz G, Calderone V, Gentili M, Sattler M, Plewka J, Taube M, Kozak M, Holak T, Dömling A, Groves M
|
| RgGuinier |
3.4 |
nm |
| Dmax |
10.5 |
nm |
| VolumePorod |
211 |
nm3 |
|
|
UniProt ID: Q6VVP7 (1-313) L-lactate dehydrogenase
|
|
|

|
| Sample: |
L-lactate dehydrogenase tetramer, 141 kDa Plasmodium falciparum protein
|
| Buffer: |
100 mM Na-phosphate buffer, 400 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at Xenocs Xeuss 2.0 with MetalJet, Department of Macromolecular Physics, Faculty of Physics, Adam Mickiewicz University on 2019 Jul 3
|
A fragment-based approach identifies an allosteric pocket that impacts malate dehydrogenase activity
Communications Biology 4(1) (2021)
Reyes Romero A, Lunev S, Popowicz G, Calderone V, Gentili M, Sattler M, Plewka J, Taube M, Kozak M, Holak T, Dömling A, Groves M
|
| RgGuinier |
3.4 |
nm |
| Dmax |
10.5 |
nm |
| VolumePorod |
244 |
nm3 |
|
|
UniProt ID: Q6VVP7 (1-313) L-lactate dehydrogenase
|
|
|
|
| Sample: |
L-lactate dehydrogenase tetramer, 141 kDa Plasmodium falciparum protein
|
| Buffer: |
100 mM Na-phosphate buffer, 400 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at Xenocs Xeuss 2.0 with MetalJet, Department of Macromolecular Physics, Faculty of Physics, Adam Mickiewicz University on 2019 Jul 3
|
A fragment-based approach identifies an allosteric pocket that impacts malate dehydrogenase activity
Communications Biology 4(1) (2021)
Reyes Romero A, Lunev S, Popowicz G, Calderone V, Gentili M, Sattler M, Plewka J, Taube M, Kozak M, Holak T, Dömling A, Groves M
|
| RgGuinier |
3.6 |
nm |
| Dmax |
11.4 |
nm |
| VolumePorod |
223 |
nm3 |
|
|
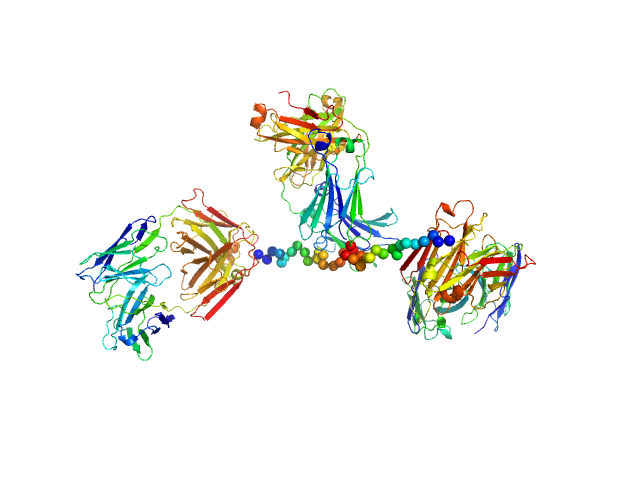
UniProt ID: None (None-None) Anti-prion protein monoclonal IgG2a 6D11
|
|
|
|
| Sample: |
Anti-prion protein monoclonal IgG2a 6D11 monomer, 145 kDa Mus musculus protein
|
| Buffer: |
phosphate buffered saline, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Dec 15
|
Ligands binding to the prion protein induce its proteolytic release with therapeutic potential in neurodegenerative proteinopathies.
Sci Adv 7(48):eabj1826 (2021)
Linsenmeier L, Mohammadi B, Shafiq M, Frontzek K, Bär J, Shrivastava AN, Damme M, Song F, Schwarz A, Da Vela S, Massignan T, Jung S, Correia A, Schmitz M, Puig B, Hornemann S, Zerr I, Tatzelt J, Biasini E, Saftig P, Schweizer M, Svergun D, Amin L, Mazzola F, Varani L, Thapa S, Gilch S, Schätzl H, Harris DA, Triller A, Mikhaylova M, Aguzzi A, Altmeppen HC, Glatzel M
|
| RgGuinier |
5.1 |
nm |
| Dmax |
17.4 |
nm |
| VolumePorod |
330 |
nm3 |
|
|
UniProt ID: P04925 (23-230) Major prion protein
UniProt ID: None (None-None) Anti-prion protein monoclonal IgG2a 6D11
|
|
|
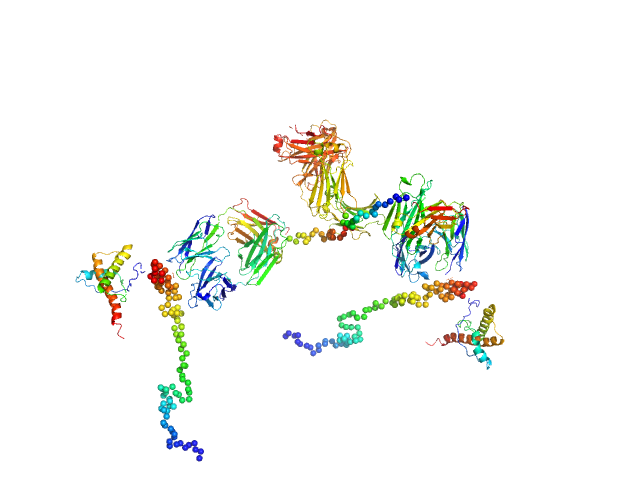
|
| Sample: |
Major prion protein monomer, 23 kDa Mus musculus protein
Anti-prion protein monoclonal IgG2a 6D11 monomer, 145 kDa Mus musculus protein
|
| Buffer: |
phosphate buffered saline, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Dec 15
|
Ligands binding to the prion protein induce its proteolytic release with therapeutic potential in neurodegenerative proteinopathies.
Sci Adv 7(48):eabj1826 (2021)
Linsenmeier L, Mohammadi B, Shafiq M, Frontzek K, Bär J, Shrivastava AN, Damme M, Song F, Schwarz A, Da Vela S, Massignan T, Jung S, Correia A, Schmitz M, Puig B, Hornemann S, Zerr I, Tatzelt J, Biasini E, Saftig P, Schweizer M, Svergun D, Amin L, Mazzola F, Varani L, Thapa S, Gilch S, Schätzl H, Harris DA, Triller A, Mikhaylova M, Aguzzi A, Altmeppen HC, Glatzel M
|
| RgGuinier |
8.1 |
nm |
| Dmax |
24.8 |
nm |
| VolumePorod |
710 |
nm3 |
|
|