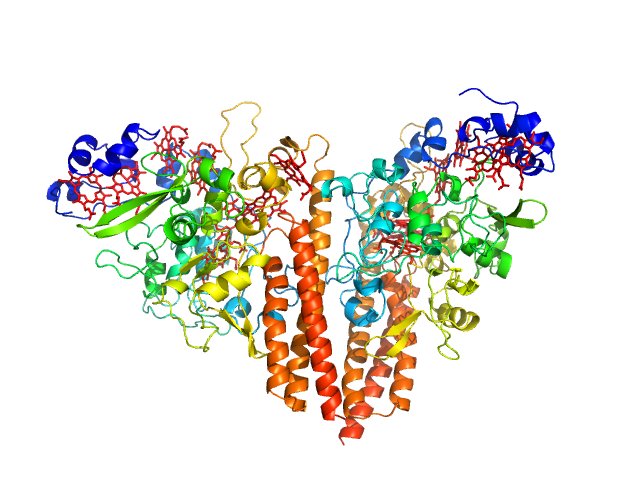
UniProt ID: L0DSL2 (29-553) Cytochrome c-552
|
|
|
|
| Sample: |
Cytochrome c-552 hexamer, 356 kDa Thioalkalivibrio nitratireducens (strain … protein
|
| Buffer: |
Tris-borate buffer, pH: 8.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2005 May 8
|
Isolation and oligomeric composition of cytochrome c nitrite reductase from the haloalkaliphilic bacterium Thioalkalivibrio nitratireducens.
Biochemistry (Mosc) 73(2):164-70 (2008)
Tikhonova TV, Slutskaya ES, Filimonenkov AA, Boyko KM, Kleimenov SY, Konarev PV, Polyakov KM, Svergun DI, Trofimov AA, Khomenkov VG, Zvyagilskaya RA, Popov VO
|
|
|
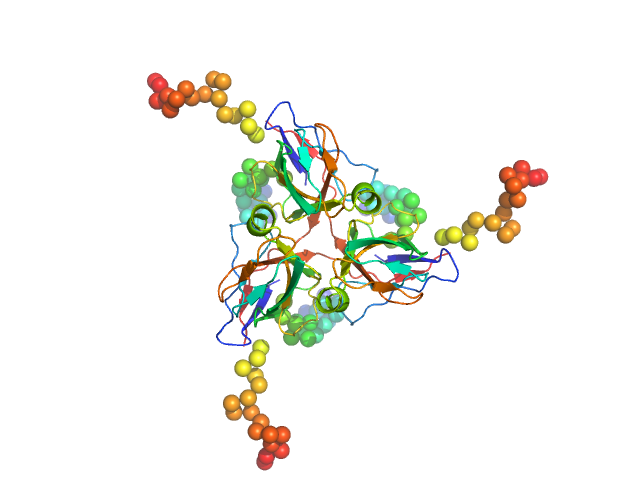
UniProt ID: P33316 (1-252) Deoxyuridine 5'-triphosphate nucleotidohydrolase, mitochondrial
|
|
|
|
| Sample: |
Deoxyuridine 5'-triphosphate nucleotidohydrolase, mitochondrial trimer, 80 kDa Homo sapiens protein
|
| Buffer: |
20 mM citrate buffer, 1 mM DTT, 0.1 mM PMSF and 1 mM MgCl2, pH: 5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2005 Feb 1
|
Molecular shape and prominent role of β-strand swapping in organization of dUTPase oligomers
FEBS Letters 583(5):865-871 (2009)
Takács E, Barabás O, Petoukhov M, Svergun D, Vértessy B
|
| RgGuinier |
2.6 |
nm |
| Dmax |
7.0 |
nm |
| VolumePorod |
101 |
nm3 |
|
|
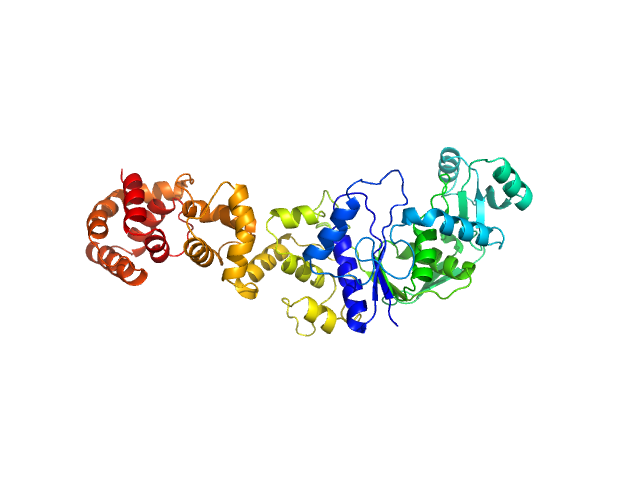
UniProt ID: P9WFV9 (1-490) Glutamate--tRNA ligase
|
|
|
|
| Sample: |
Glutamate--tRNA ligase dimer, 108 kDa Mycobacterium tuberculosis (strain … protein
|
| Buffer: |
35 mM HEPES⁄NaOH, pH: 6.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2007 Feb 19
|
Kinetic and mechanistic characterization of Mycobacterium tuberculosis glutamyl-tRNA synthetase and determination of its oligomeric structure in solution
FEBS Journal 276(5):1398-1417 (2009)
Paravisi S, Fumagalli G, Riva M, Morandi P, Morosi R, Konarev P, Petoukhov M, Bernier S, Chênevert R, Svergun D, Curti B, Vanoni M
|
| RgGuinier |
3.5 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
123 |
nm3 |
|
|
UniProt ID: P14210 (38-209) Hepatocyte growth factor
UniProt ID: P08581 (25-562) Hepatocyte growth factor receptor
|
|
|

|
| Sample: |
Hepatocyte growth factor dimer, 39 kDa Homo sapiens protein
Hepatocyte growth factor receptor dimer, 122 kDa Homo sapiens protein
|
| Buffer: |
Phosphate-buffered saline containing EDTA, surfactant P20 and 5 M NaCl, pH: 6 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2007 Jun 26
|
Engineering the NK1 Fragment of Hepatocyte Growth Factor/Scatter Factor as a MET Receptor Antagonist
Journal of Molecular Biology 377(3):616-622 (2008)
Youles M, Holmes O, Petoukhov M, Nessen M, Stivala S, Svergun D, Gherardi E
|
| RgGuinier |
5.3 |
nm |
| Dmax |
20.0 |
nm |
| VolumePorod |
290 |
nm3 |
|
|
UniProt ID: Q9VDG5 (442-697) AT07459p
|
|
|
|
| Sample: |
AT07459p monomer, 30 kDa Drosophila melanogaster protein
|
| Buffer: |
20 mM Tris, 5 mM β-mercaptoethanol, 200 mM NaCl, 1% v/v glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at Xenocs Xeuss, Institute of Physics, University of São Paulo on 2018 Mar 8
|
Molecular insights on CALX-CBD12 inter-domain dynamics from MD simulations, RDCs and SAXS.
Biophys J (2021)
de Souza Degenhardt MF, Vitale PAM, Abiko LA, Zacharias M, Sattler M, Oliveira CLP, Salinas RK
|
| RgGuinier |
2.5 |
nm |
| Dmax |
8.7 |
nm |
| VolumePorod |
41 |
nm3 |
|
|
UniProt ID: Q9VDG5 (442-697) AT07459p
|
|
|
|
| Sample: |
AT07459p monomer, 30 kDa Drosophila melanogaster protein
|
| Buffer: |
20 mM Tris, 5 mM β-mercaptoethanol, 200 mM NaCl, 1% v/v glycerol, 0.8 mM CaCl2, pH: 7.4 |
| Experiment: |
SAXS
data collected at Xenocs Xeuss, Institute of Physics, University of São Paulo on 2018 Apr 13
|
Molecular insights on CALX-CBD12 inter-domain dynamics from MD simulations, RDCs and SAXS.
Biophys J (2021)
de Souza Degenhardt MF, Vitale PAM, Abiko LA, Zacharias M, Sattler M, Oliveira CLP, Salinas RK
|
| RgGuinier |
2.7 |
nm |
| Dmax |
9.3 |
nm |
| VolumePorod |
40 |
nm3 |
|
|
UniProt ID: E9PW06 (25-621) Neurofascin
|
|
|
|
| Sample: |
Neurofascin dimer, 139 kDa Mus musculus protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Dec 16
|
Structural insights into the contactin 1 – neurofascin 155 adhesion complex
Nature Communications 13(1) (2022)
Chataigner L, Gogou C, den Boer M, Frias C, Thies-Weesie D, Granneman J, Heck A, Meijer D, Janssen B
|
| RgGuinier |
4.7 |
nm |
| Dmax |
18.0 |
nm |
| VolumePorod |
355 |
nm3 |
|
|
UniProt ID: E9PW06 (25-621) Neurofascin
|
|
|
|
| Sample: |
Neurofascin dimer, 139 kDa Mus musculus protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Dec 16
|
Structural insights into the contactin 1 – neurofascin 155 adhesion complex
Nature Communications 13(1) (2022)
Chataigner L, Gogou C, den Boer M, Frias C, Thies-Weesie D, Granneman J, Heck A, Meijer D, Janssen B
|
| RgGuinier |
5.0 |
nm |
| Dmax |
21.0 |
nm |
| VolumePorod |
450 |
nm3 |
|
|
UniProt ID: E9PW06 (25-621) Neurofascin
|
|
|
|
| Sample: |
Neurofascin dimer, 139 kDa Mus musculus protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Dec 16
|
Structural insights into the contactin 1 – neurofascin 155 adhesion complex
Nature Communications 13(1) (2022)
Chataigner L, Gogou C, den Boer M, Frias C, Thies-Weesie D, Granneman J, Heck A, Meijer D, Janssen B
|
| RgGuinier |
4.5 |
nm |
| Dmax |
17.0 |
nm |
| VolumePorod |
290 |
nm3 |
|
|
UniProt ID: E9PW06 (25-621) Neurofascin
|
|
|
|
| Sample: |
Neurofascin dimer, 139 kDa Mus musculus protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Dec 16
|
Structural insights into the contactin 1 – neurofascin 155 adhesion complex
Nature Communications 13(1) (2022)
Chataigner L, Gogou C, den Boer M, Frias C, Thies-Weesie D, Granneman J, Heck A, Meijer D, Janssen B
|
| RgGuinier |
4.3 |
nm |
| Dmax |
17.0 |
nm |
| VolumePorod |
240 |
nm3 |
|
|