UniProt ID: P12960 (21-996) Contactin-1 I433V
|
|
|
|
| Sample: |
Contactin-1 I433V dimer, 220 kDa Mus musculus protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Dec 16
|
Structural insights into the contactin 1 – neurofascin 155 adhesion complex
Nature Communications 13(1) (2022)
Chataigner L, Gogou C, den Boer M, Frias C, Thies-Weesie D, Granneman J, Heck A, Meijer D, Janssen B
|
| RgGuinier |
6.8 |
nm |
| Dmax |
27.0 |
nm |
| VolumePorod |
275 |
nm3 |
|
|
UniProt ID: P12960 (21-996) Contactin-1 I433V
|
|
|
|
| Sample: |
Contactin-1 I433V dimer, 220 kDa Mus musculus protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Dec 16
|
Structural insights into the contactin 1 – neurofascin 155 adhesion complex
Nature Communications 13(1) (2022)
Chataigner L, Gogou C, den Boer M, Frias C, Thies-Weesie D, Granneman J, Heck A, Meijer D, Janssen B
|
| RgGuinier |
6.8 |
nm |
| Dmax |
27.0 |
nm |
| VolumePorod |
270 |
nm3 |
|
|
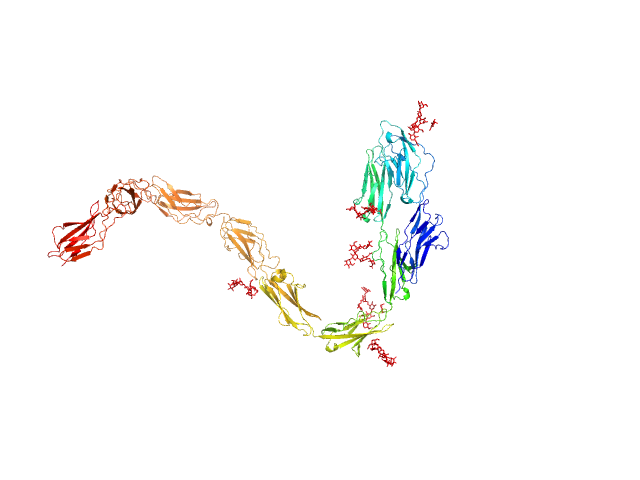
UniProt ID: P12960 (21-996) Contactin-1 I433V
|
|
|
|
| Sample: |
Contactin-1 I433V dimer, 220 kDa Mus musculus protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Dec 16
|
Structural insights into the contactin 1 – neurofascin 155 adhesion complex
Nature Communications 13(1) (2022)
Chataigner L, Gogou C, den Boer M, Frias C, Thies-Weesie D, Granneman J, Heck A, Meijer D, Janssen B
|
| RgGuinier |
6.8 |
nm |
| Dmax |
19.5 |
nm |
| VolumePorod |
254 |
nm3 |
|
|
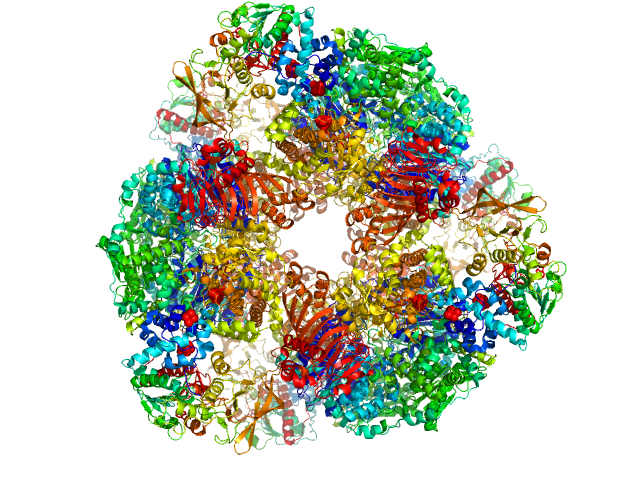
UniProt ID: Q05755 (1-1508) Glutamate synthase [NADPH] large chain
UniProt ID: Q05756 (27-482) Glutamate synthase [NADPH] small chain
|
|
|
|
| Sample: |
Glutamate synthase [NADPH] large chain hexamer, 968 kDa Azospirillum brasilense protein
Glutamate synthase [NADPH] small chain hexamer, 296 kDa Azospirillum brasilense protein
|
| Buffer: |
25 mM Hepes/KOH, 1 mM EDTA, 1 mM dithiothreitol, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2007 Feb 19
|
The Subnanometer Resolution Structure of the Glutamate Synthase 1.2-MDa Hexamer by Cryoelectron Microscopy and Its Oligomerization Behavior in Solution
Journal of Biological Chemistry 283(13):8237-8249 (2008)
Cottevieille M, Larquet E, Jonic S, Petoukhov M, Caprini G, Paravisi S, Svergun D, Vanoni M, Boisset N
|
| RgGuinier |
7.7 |
nm |
| Dmax |
22.4 |
nm |
|
|
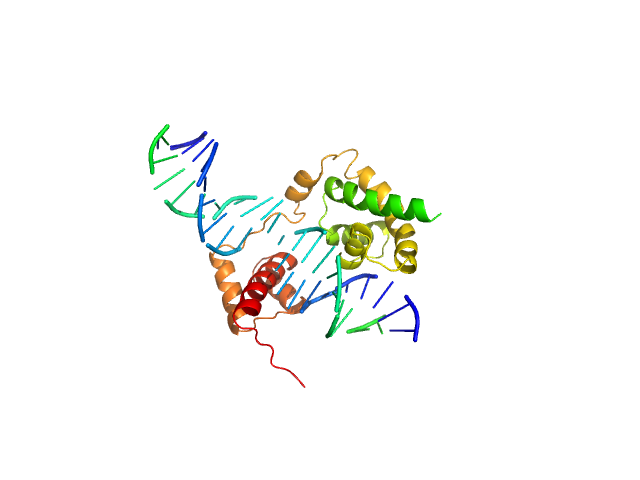
UniProt ID: P20265 (254-420) POU domain, class 3, transcription factor 2
UniProt ID: None (None-None) Rat CRH DNA
|
|
|
|
| Sample: |
POU domain, class 3, transcription factor 2 monomer, 19 kDa Homo sapiens protein
Rat CRH DNA monomer, 15 kDa Rattus norvegicus DNA
|
| Buffer: |
50 mM Tris, 0.4 M NaCl, 2% glycerol, 2 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2003 Oct 9
|
Fine-tuning of intrinsic N-Oct-3 POU domain allostery by regulatory DNA targets
Nucleic Acids Research 35(13):4420-4432 (2007)
Alazard R, Mourey L, Ebel C, Konarev P, Petoukhov M, Svergun D, Erard M
|
| RgGuinier |
2.9 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
41 |
nm3 |
|
|
UniProt ID: P20265 (254-420) POU domain, class 3, transcription factor 2
UniProt ID: None (None-None) Human DR-alpha DNA
|
|
|
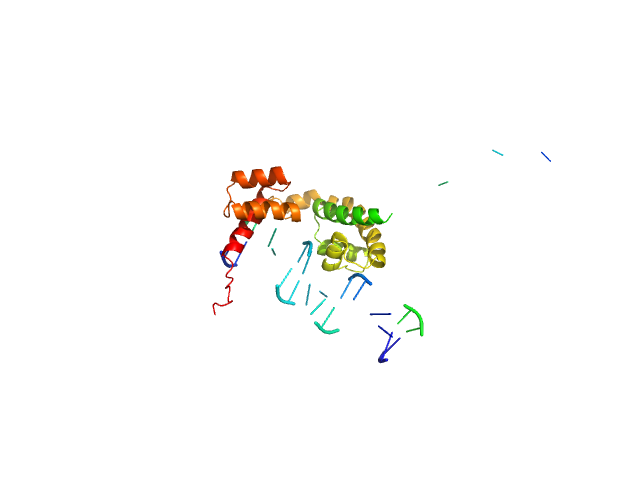
|
| Sample: |
POU domain, class 3, transcription factor 2 monomer, 19 kDa Homo sapiens protein
Human DR-alpha DNA monomer, 15 kDa Homo sapiens DNA
|
| Buffer: |
50 mM Tris, 0.4 M NaCl, 2% glycerol, 2 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2004 May 26
|
Fine-tuning of intrinsic N-Oct-3 POU domain allostery by regulatory DNA targets
Nucleic Acids Research 35(13):4420-4432 (2007)
Alazard R, Mourey L, Ebel C, Konarev P, Petoukhov M, Svergun D, Erard M
|
| RgGuinier |
2.9 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
44 |
nm3 |
|
|
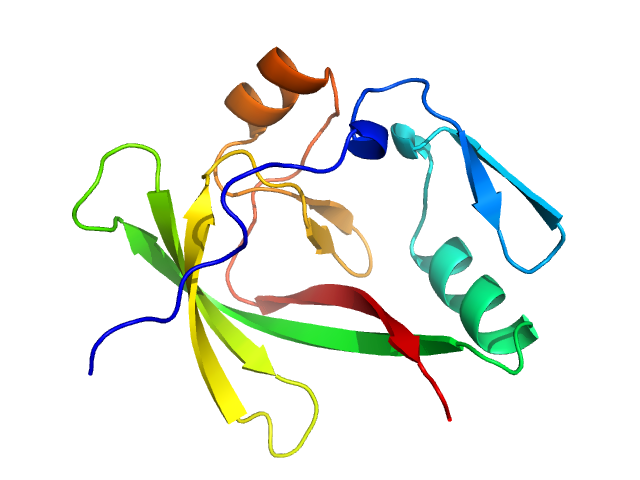
UniProt ID: P54253 (567-689) Ataxin-1
|
|
|
|
| Sample: |
Ataxin-1 monomer, 14 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris-HCl, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2010 Oct 21
|
Self-assembly and conformational heterogeneity of the AXH domain of ataxin-1: an unusual example of a chameleon fold.
Biophys J 104(6):1304-13 (2013)
de Chiara C, Rees M, Menon RP, Pauwels K, Lawrence C, Konarev PV, Svergun DI, Martin SR, Chen YW, Pastore A
|
|
|
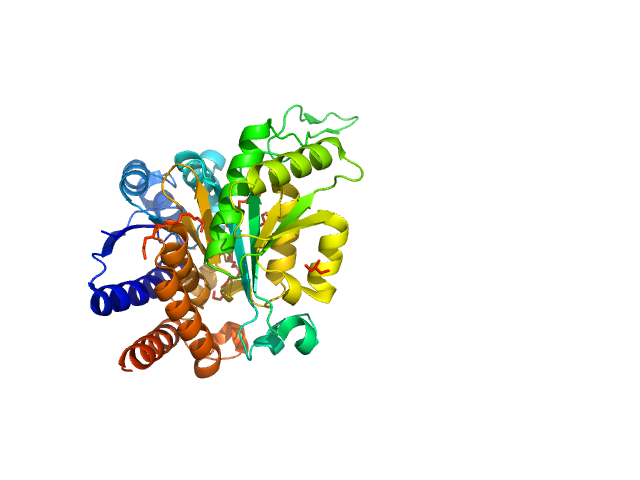
UniProt ID: P61495 (1-345) 3-isopropylmalate dehydrogenase
|
|
|
|
| Sample: |
3-isopropylmalate dehydrogenase dimer, 73 kDa Thermus thermophilus protein
|
| Buffer: |
25 mM MOPS/NaOH, pH: 7.6 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Dec 3
|
Glutamate 270 plays an essential role in K(+)-activation and domain closure of Thermus thermophilus isopropylmalate dehydrogenase.
FEBS Lett 589(2):240-5 (2015)
Gráczer É, Palló A, Oláh J, Szimler T, Konarev PV, Svergun DI, Merli A, Závodszky P, Weiss MS, Vas M
|
|
|
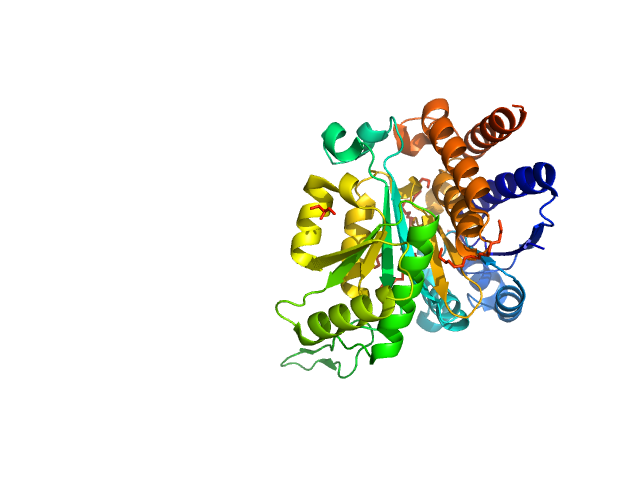
UniProt ID: P61495 (1-345) 3-isopropylmalate dehydrogenase
|
|
|
|
| Sample: |
3-isopropylmalate dehydrogenase dimer, 73 kDa Thermus thermophilus protein
|
| Buffer: |
25 mM MOPS/NaOH, pH: 7.6 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2012 Nov 16
|
Dual Role of the Active Site Residues of Thermus thermophilus 3-Isopropylmalate Dehydrogenase: Chemical Catalysis and Domain Closure.
Biochemistry 55(3):560-74 (2016)
Gráczer É, Szimler T, Garamszegi A, Konarev PV, Lábas A, Oláh J, Palló A, Svergun DI, Merli A, Závodszky P, Weiss MS, Vas M
|
|
|
UniProt ID: Q8EG35 (35-333) Extracelllular iron oxide respiratory system periplasmic decaheme cytochrome c component MtrA
UniProt ID: Q8EG34 (26-671) Extracellular iron oxide respiratory system surface decaheme cytochrome c component MtrC
UniProt ID: Q8CVD4 (22-697) Extracellular iron oxide respiratory system outer membrane component MtrB
|
|
|

|
| Sample: |
Extracelllular iron oxide respiratory system periplasmic decaheme cytochrome c component MtrA monomer, 39 kDa Shewanella oneidensis (strain … protein
Extracellular iron oxide respiratory system surface decaheme cytochrome c component MtrC monomer, 77 kDa Shewanella oneidensis (strain … protein
Extracellular iron oxide respiratory system outer membrane component MtrB monomer, 75 kDa Shewanella oneidensis (strain … protein
|
| Buffer: |
20 mM HEPES, 100 mM NaCl, 2.8 mM Fos-choline 12, 13% D2O, pH: 7.8 |
| Experiment: |
SANS
data collected at D22, Institut Laue-Langevin (ILL) on 2020 Jan 13
|
Bespoke Biomolecular Wires for Transmembrane Electron Transfer: Spontaneous Assembly of a Functionalized Multiheme Electron Conduit
Frontiers in Microbiology 12 (2021)
Piper S, Edwards M, van Wonderen J, Casadevall C, Martel A, Jeuken L, Reisner E, Clarke T, Butt J
|
| RgGuinier |
4.7 |
nm |
| Dmax |
16.6 |
nm |
| VolumePorod |
154 |
nm3 |
|
|