UniProt ID: T2HVF7 (None-None) Cyclopentadecanone 1,2-monooxygenase
|
|
|
|
| Sample: |
Cyclopentadecanone 1,2-monooxygenase monomer, 68 kDa Pseudomonas sp. HI-70 protein
|
| Buffer: |
50mM Tris 2mM TCEP 5mM NADP+ 1mM cyclopentadecanon, pH: 8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2012 Oct 2
|
The role of conformational flexibility in Baeyer-Villiger monooxygenase catalysis and structure.
Biochim Biophys Acta 1864(12):1641-1648 (2016)
Yachnin BJ, Lau PCK, Berghuis AM
|
| RgGuinier |
2.6 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
110 |
nm3 |
|
|
UniProt ID: T2HVF7 (None-None) Cyclopentadecanone 1,2-monooxygenase
|
|
|
|
| Sample: |
Cyclopentadecanone 1,2-monooxygenase monomer, 68 kDa Pseudomonas sp. HI-70 protein
|
| Buffer: |
50mM Tris mM TCEP 5mM NADP+ 1mM ω-pentadecalactone, pH: 8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2012 Oct 2
|
The role of conformational flexibility in Baeyer-Villiger monooxygenase catalysis and structure.
Biochim Biophys Acta 1864(12):1641-1648 (2016)
Yachnin BJ, Lau PCK, Berghuis AM
|
| RgGuinier |
2.6 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
100 |
nm3 |
|
|
UniProt ID: Q9VYW2 (509-716) FI10506p
|
|
|
|
| Sample: |
FI10506p monomer, 23 kDa Drosophila melanogaster protein
|
| Buffer: |
20mM Tris/HCl 150mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2015 Jul 21
|
Intrinsic Disorder of the C-Terminal Domain of Drosophila Methoprene-Tolerant Protein.
PLoS One 11(9):e0162950 (2016)
Kolonko M, Ożga K, Hołubowicz R, Taube M, Kozak M, Ożyhar A, Greb-Markiewicz B
|
| RgGuinier |
5.1 |
nm |
| Dmax |
22.0 |
nm |
|
|
UniProt ID: P0A9N0 (1-85) Phosphocarrier protein NPr
UniProt ID: P37177 (170-424) Phosphoenolpyruvate-protein phosphotransferase PtsP
|
|
|
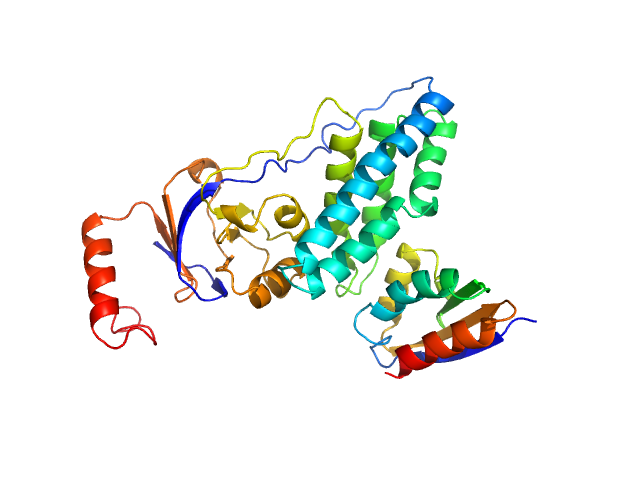
|
| Sample: |
Phosphocarrier protein NPr monomer, 9 kDa Escherichia coli protein
Phosphoenolpyruvate-protein phosphotransferase PtsP monomer, 28 kDa Escherichia coli protein
|
| Buffer: |
10 mM Tris 150 mM NaCl 0.5 mM EDTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-1000, NIDDK, NIH on 2015 Jan 30
|
Structure of the NPr:EINNtr Complex: Mechanism for Specificity in Paralogous Phosphotransferase Systems.
Structure 24(12):2127-2137 (2016)
Strickland M, Stanley AM, Wang G, Botos I, Schwieters CD, Buchanan SK, Peterkofsky A, Tjandra N
|
| RgGuinier |
2.3 |
nm |
| Dmax |
8.2 |
nm |
| VolumePorod |
52 |
nm3 |
|
|
UniProt ID: O60885 (1-477) Bromodomain-containing protein 4
|
|
|

|
| Sample: |
Bromodomain-containing protein 4 monomer, 56 kDa Homo sapiens protein
|
| Buffer: |
20mM Hepes, 100mM NaCl, 1mM Tris(2-carboxyethyl)phosphine hydrochloride, pH: 7.4 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2014 Sep 12
|
Potent and selective bivalent inhibitors of BET bromodomains.
Nat Chem Biol 12(12):1097-1104 (2016)
Waring MJ, Chen H, Rabow AA, Walker G, Bobby R, Boiko S, Bradbury RH, Callis R, Clark E, Dale I, Daniels DL, Dulak A, Flavell L, Holdgate G, Jowitt TA, Kikhney A, McAlister M, Méndez J, Ogg D, Patel J, Petteruti P, Robb GR, Robers MB, Saif S, Stratton N, Svergun DI, Wang W, Whittaker D, Wilson DM, Yao Y
|
| RgGuinier |
7.0 |
nm |
| Dmax |
27.0 |
nm |
|
|
UniProt ID: O60885 (1-477) Bromodomain-containing protein 4
|
|
|
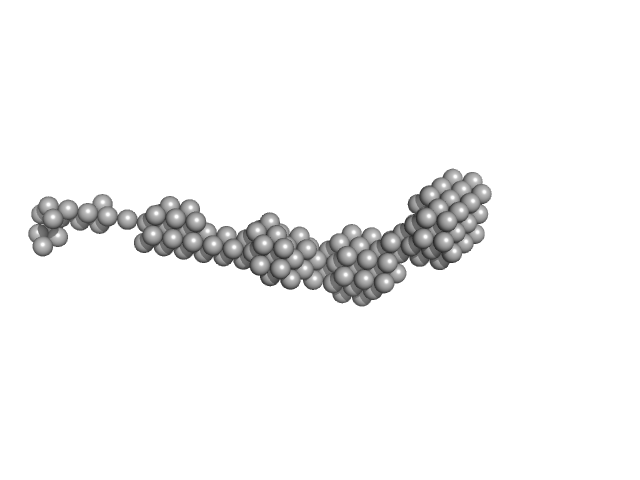
|
| Sample: |
Bromodomain-containing protein 4 monomer, 56 kDa Homo sapiens protein
|
| Buffer: |
20mM Hepes, 100mM NaCl, 1mM Tris(2-carboxyethyl)phosphine hydrochloride, pH: 7.4 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2014 Sep 12
|
Potent and selective bivalent inhibitors of BET bromodomains.
Nat Chem Biol 12(12):1097-1104 (2016)
Waring MJ, Chen H, Rabow AA, Walker G, Bobby R, Boiko S, Bradbury RH, Callis R, Clark E, Dale I, Daniels DL, Dulak A, Flavell L, Holdgate G, Jowitt TA, Kikhney A, McAlister M, Méndez J, Ogg D, Patel J, Petteruti P, Robb GR, Robers MB, Saif S, Stratton N, Svergun DI, Wang W, Whittaker D, Wilson DM, Yao Y
|
| RgGuinier |
6.2 |
nm |
| Dmax |
27.5 |
nm |
|
|
UniProt ID: O60885 (1-477) Bromodomain-containing protein 4
|
|
|
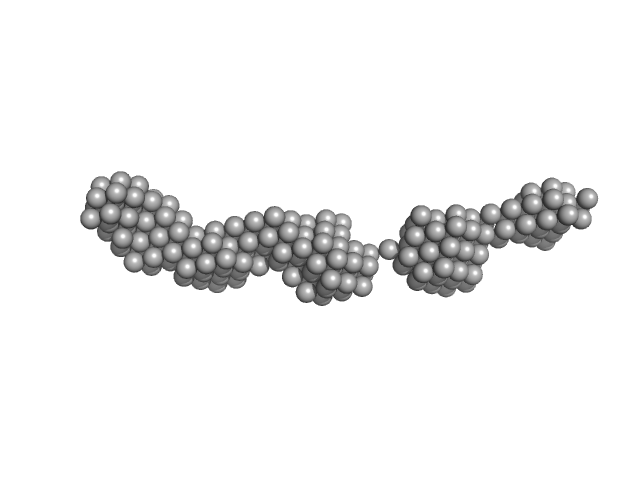
|
| Sample: |
Bromodomain-containing protein 4 monomer, 56 kDa Homo sapiens protein
|
| Buffer: |
20mM Hepes, 100mM NaCl, 1mM Tris(2-carboxyethyl)phosphine hydrochloride, pH: 7.4 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2014 Sep 12
|
Potent and selective bivalent inhibitors of BET bromodomains.
Nat Chem Biol 12(12):1097-1104 (2016)
Waring MJ, Chen H, Rabow AA, Walker G, Bobby R, Boiko S, Bradbury RH, Callis R, Clark E, Dale I, Daniels DL, Dulak A, Flavell L, Holdgate G, Jowitt TA, Kikhney A, McAlister M, Méndez J, Ogg D, Patel J, Petteruti P, Robb GR, Robers MB, Saif S, Stratton N, Svergun DI, Wang W, Whittaker D, Wilson DM, Yao Y
|
| RgGuinier |
6.3 |
nm |
| Dmax |
24.5 |
nm |
|
|
UniProt ID: O60885 (1-477) Bromodomain-containing protein 4
|
|
|
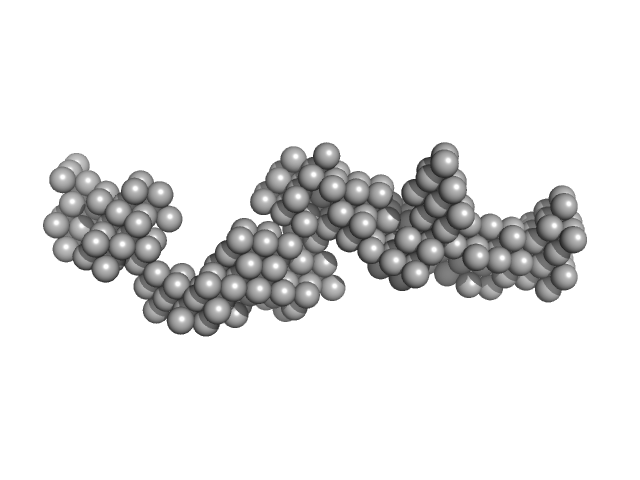
|
| Sample: |
Bromodomain-containing protein 4 monomer, 56 kDa Homo sapiens protein
|
| Buffer: |
20mM Hepes, 100mM NaCl, 1mM Tris(2-carboxyethyl)phosphine hydrochloride, pH: 7.4 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2015 Feb 8
|
Potent and selective bivalent inhibitors of BET bromodomains.
Nat Chem Biol 12(12):1097-1104 (2016)
Waring MJ, Chen H, Rabow AA, Walker G, Bobby R, Boiko S, Bradbury RH, Callis R, Clark E, Dale I, Daniels DL, Dulak A, Flavell L, Holdgate G, Jowitt TA, Kikhney A, McAlister M, Méndez J, Ogg D, Patel J, Petteruti P, Robb GR, Robers MB, Saif S, Stratton N, Svergun DI, Wang W, Whittaker D, Wilson DM, Yao Y
|
| RgGuinier |
5.1 |
nm |
| Dmax |
24.3 |
nm |
|
|
UniProt ID: O60885 (1-477) Bromodomain-containing protein 4
|
|
|

|
| Sample: |
Bromodomain-containing protein 4 monomer, 56 kDa Homo sapiens protein
|
| Buffer: |
20mM Hepes, 100mM NaCl, 1mM Tris(2-carboxyethyl)phosphine hydrochloride, pH: 7.4 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2015 Feb 8
|
Potent and selective bivalent inhibitors of BET bromodomains.
Nat Chem Biol 12(12):1097-1104 (2016)
Waring MJ, Chen H, Rabow AA, Walker G, Bobby R, Boiko S, Bradbury RH, Callis R, Clark E, Dale I, Daniels DL, Dulak A, Flavell L, Holdgate G, Jowitt TA, Kikhney A, McAlister M, Méndez J, Ogg D, Patel J, Petteruti P, Robb GR, Robers MB, Saif S, Stratton N, Svergun DI, Wang W, Whittaker D, Wilson DM, Yao Y
|
| RgGuinier |
4.4 |
nm |
| Dmax |
17.3 |
nm |
|
|
UniProt ID: P53927 (81-220) Ribosome biogenesis protein 15
|
|
|
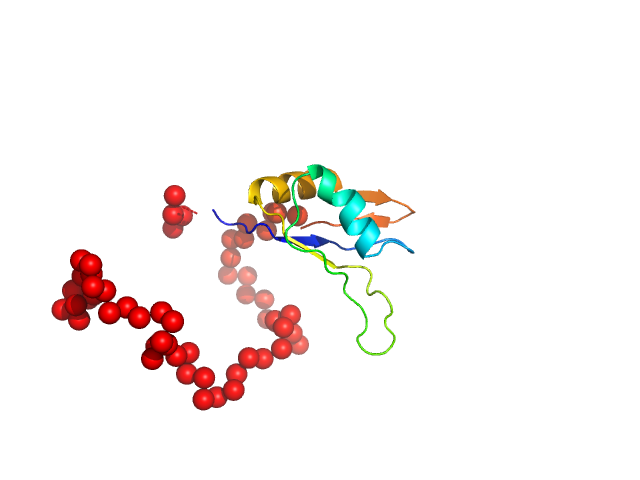
|
| Sample: |
Ribosome biogenesis protein 15 monomer, 17 kDa Saccharomyces cerevisiae protein
|
| Buffer: |
25 mM HEPES, 500 mM NaCl, 2 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2013 Apr 25
|
Structural analysis reveals the flexible C-terminus of Nop15 undergoes rearrangement to recognize a pre-ribosomal RNA folding intermediate.
Nucleic Acids Res 45(5):2829-2837 (2017)
Zhang J, Gonzalez LE, Hall TMT
|
| RgGuinier |
2.4 |
nm |
| Dmax |
10.3 |
nm |
| VolumePorod |
38 |
nm3 |
|
|