UniProt ID: P42641 (1-390) GTPase ObgE/CgtA
|
|
|
|
| Sample: |
GTPase ObgE/CgtA monomer, 44 kDa Escherichia coli protein
|
| Buffer: |
20 mM Hepes , 300 mM NaCl, 250 mM imidazole, 5 mM MgCl2, 2 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2015 Jun 17
|
Structural and biochemical analysis of Escherichia coli ObgE, a central regulator of bacterial persistence.
J Biol Chem 292(14):5871-5883 (2017)
Gkekas S, Singh RK, Shkumatov AV, Messens J, Fauvart M, Verstraeten N, Michiels J, Versées W
|
| RgGuinier |
3.7 |
nm |
| Dmax |
18.1 |
nm |
| VolumePorod |
102 |
nm3 |
|
|
UniProt ID: P42641 (1-390) GTPase ObgE/CgtA
|
|
|

|
| Sample: |
GTPase ObgE/CgtA monomer, 44 kDa Escherichia coli protein
|
| Buffer: |
20 mM Hepes, 300 mM NaCl, 250 mM imidazole, 5 mM MgCl2, 2 mM DTT, 400 µM GppNHp, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2015 Jun 17
|
Structural and biochemical analysis of Escherichia coli ObgE, a central regulator of bacterial persistence.
J Biol Chem 292(14):5871-5883 (2017)
Gkekas S, Singh RK, Shkumatov AV, Messens J, Fauvart M, Verstraeten N, Michiels J, Versées W
|
| RgGuinier |
3.7 |
nm |
| Dmax |
14.5 |
nm |
| VolumePorod |
90 |
nm3 |
|
|
UniProt ID: P42641 (1-340) GTPase ObgE/CgtA
|
|
|
|
| Sample: |
GTPase ObgE/CgtA monomer, 39 kDa Escherichia coli protein
|
| Buffer: |
20 mM Hepes , 300 mM NaCl, 250 mM imidazole, 5 mM MgCl2, 2 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2015 Mar 12
|
Structural and biochemical analysis of Escherichia coli ObgE, a central regulator of bacterial persistence.
J Biol Chem 292(14):5871-5883 (2017)
Gkekas S, Singh RK, Shkumatov AV, Messens J, Fauvart M, Verstraeten N, Michiels J, Versées W
|
| RgGuinier |
3.1 |
nm |
| Dmax |
11.6 |
nm |
| VolumePorod |
70 |
nm3 |
|
|
UniProt ID: P42641 (1-340) GTPase ObgE/CgtA
|
|
|

|
| Sample: |
GTPase ObgE/CgtA monomer, 39 kDa Escherichia coli protein
|
| Buffer: |
20 mM Hepes, 300 mM NaCl, 250 mM imidazole, 5 mM MgCl2, 2 mM DTT, 400 µM GppNHp, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2015 Mar 12
|
Structural and biochemical analysis of Escherichia coli ObgE, a central regulator of bacterial persistence.
J Biol Chem 292(14):5871-5883 (2017)
Gkekas S, Singh RK, Shkumatov AV, Messens J, Fauvart M, Verstraeten N, Michiels J, Versées W
|
| RgGuinier |
3.1 |
nm |
| Dmax |
11.4 |
nm |
| VolumePorod |
72 |
nm3 |
|
|
UniProt ID: C2LPE2 (22-243) Suppressor of Copper Sensitivity C protein (mutant)
|
|
|
|
| Sample: |
Suppressor of Copper Sensitivity C protein (mutant) trimer, 73 kDa Proteus mirabilis protein
|
| Buffer: |
25 mM HEPES 150mM NaCl, 1mM DTT,, pH: 7.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2016 Jul 22
|
A shape-shifting redox foldase contributes to Proteus mirabilis copper resistance.
Nat Commun 8:16065 (2017)
Furlong EJ, Lo AW, Kurth F, Premkumar L, Totsika M, Achard MES, Halili MA, Heras B, Whitten AE, Choudhury HG, Schembri MA, Martin JL
|
| RgGuinier |
4.4 |
nm |
| Dmax |
13.5 |
nm |
| VolumePorod |
108 |
nm3 |
|
|
UniProt ID: Q8NBI3 (None-None) Draxin
|
|
|
|
| Sample: |
Draxin monomer, 45 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2015 Aug 19
|
Structural Basis for Draxin-Modulated Axon Guidance and Fasciculation by Netrin-1 through DCC.
Neuron 97(6):1261-1267.e4 (2018)
Liu Y, Bhowmick T, Liu Y, Gao X, Mertens HDT, Svergun DI, Xiao J, Zhang Y, Wang JH, Meijers R
|
| RgGuinier |
4.2 |
nm |
| Dmax |
15.0 |
nm |
| VolumePorod |
87 |
nm3 |
|
|
UniProt ID: P54397 (1-357) 39 kDa FK506-binding nuclear protein
|
|
|
|
| Sample: |
39 kDa FK506-binding nuclear protein tetramer, 163 kDa Drosophila melanogaster protein
|
| Buffer: |
50 mM Na2HPO4 150 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2014 Nov 14
|
Nucleoplasmin-like domain of FKBP39 from Drosophila melanogaster forms a tetramer with partly disordered tentacle-like C-terminal segments.
Sci Rep 7:40405 (2017)
Kozłowska M, Tarczewska A, Jakób M, Bystranowska D, Taube M, Kozak M, Czarnocki-Cieciura M, Dziembowski A, Orłowski M, Tkocz K, Ożyhar A
|
| RgGuinier |
5.7 |
nm |
| Dmax |
22.0 |
nm |
| VolumePorod |
275 |
nm3 |
|
|
UniProt ID: I0RZI0 (None-None) ESX-5 type VII secretion system protein EccC5
|
|
|
|
| Sample: |
ESX-5 type VII secretion system protein EccC5 monomer, 142 kDa Mycobacterium xenopi RIVM700367 protein
|
| Buffer: |
20 mM Tris 200 mM NaCl 5%(v/v) Glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Jul 15
|
Structure of the mycobacterial ESX-5 type VII secretion system membrane complex by single-particle analysis.
Nat Microbiol 2:17047 (2017)
Beckham KS, Ciccarelli L, Bunduc CM, Mertens HD, Ummels R, Lugmayr W, Mayr J, Rettel M, Savitski MM, Svergun DI, Bitter W, Wilmanns M, Marlovits TC, Parret AH, Houben EN
|
| RgGuinier |
6.2 |
nm |
| Dmax |
23.0 |
nm |
| VolumePorod |
255 |
nm3 |
|
|
UniProt ID: P26672 (None-None) Protein A46
|
|
|
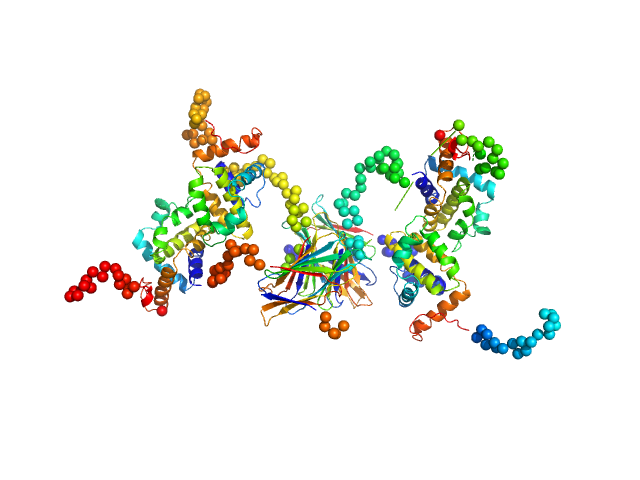
|
| Sample: |
Protein A46 tetramer, 112 kDa Vaccinia virus protein
|
| Buffer: |
20 mM Tris-HCl, 10 mM DTT, pH: 8.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2015 Jun 25
|
Vaccinia Virus Immunomodulator A46: A Lipid and Protein-Binding Scaffold for Sequestering Host TIR-Domain Proteins.
PLoS Pathog 12(12):e1006079 (2016)
Fedosyuk S, Bezerra GA, Radakovics K, Smith TK, Sammito M, Bobik N, Round A, Ten Eyck LF, Djinović-Carugo K, Usón I, Skern T
|
| RgGuinier |
4.3 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
199 |
nm3 |
|
|
UniProt ID: P26672 (1-83) Protein A46
|
|
|
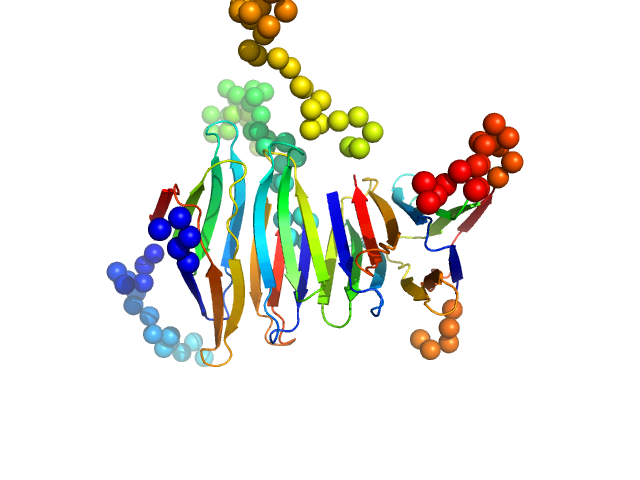
|
| Sample: |
Protein A46 tetramer, 40 kDa Vaccinia virus protein
|
| Buffer: |
20 mM Tris-HCl, 10 mM DTT, pH: 8.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2015 Jun 25
|
Vaccinia Virus Immunomodulator A46: A Lipid and Protein-Binding Scaffold for Sequestering Host TIR-Domain Proteins.
PLoS Pathog 12(12):e1006079 (2016)
Fedosyuk S, Bezerra GA, Radakovics K, Smith TK, Sammito M, Bobik N, Round A, Ten Eyck LF, Djinović-Carugo K, Usón I, Skern T
|
| RgGuinier |
2.6 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
68 |
nm3 |
|
|