UniProt ID: P69905 (3-142) Hemoglobin subunit alpha
UniProt ID: P68871 (3-147) Hemoglobin subunit beta
|
|
|
|
| Sample: |
Hemoglobin subunit alpha monomer, 15 kDa Homo sapiens protein
Hemoglobin subunit beta monomer, 16 kDa Homo sapiens protein
|
| Buffer: |
Ringer's lactate solution, pH: 6.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2006 Feb 19
|
Solution Structure of Poly(ethylene) Glycol-Conjugated Hemoglobin Revealed by Small-Angle X-Ray Scattering: Implications for a New Oxygen Therapeutic
Biophysical Journal 94(1):173-181 (2008)
Svergun D, Ekström F, Vandegriff K, Malavalli A, Baker D, Nilsson C, Winslow R
|
|
|
UniProt ID: P02790 (None-None) Hemopexin
UniProt ID: P02790 (None-None) Hemopexin
|
|
|
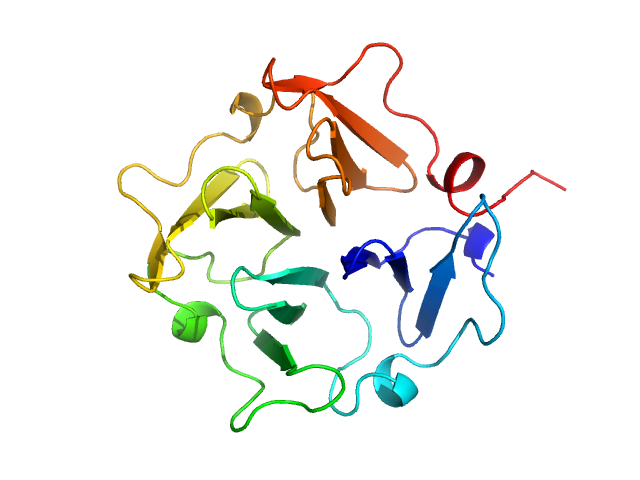
|
| Sample: |
Hemopexin monomer, 23 kDa Homo sapiens protein
Hemopexin dimer, 46 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, 150 mM NaCl, 10 mM CaCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2006 Nov 25
|
The Dimer Interface of the Membrane Type 1 Matrix Metalloproteinase Hemopexin Domain
Journal of Biological Chemistry 286(9):7587-7600 (2011)
Tochowicz A, Goettig P, Evans R, Visse R, Shitomi Y, Palmisano R, Ito N, Richter K, Maskos K, Franke D, Svergun D, Nagase H, Bode W, Itoh Y
|
| RgGuinier |
2.3 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
48 |
nm3 |
|
|
UniProt ID: Q91132 (None-None) Cobra venom factor
|
|
|
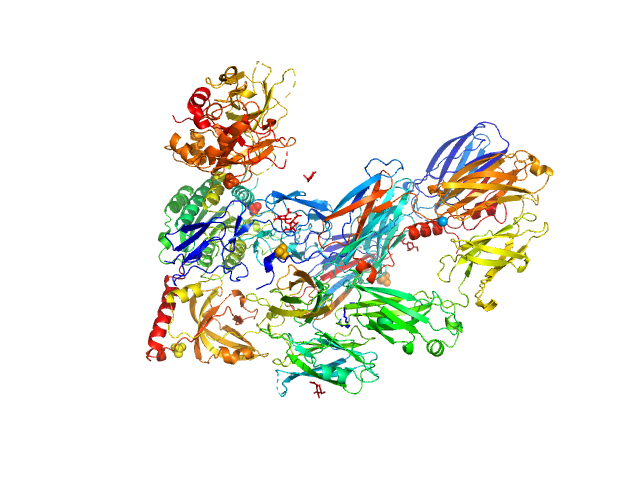
|
| Sample: |
Cobra venom factor monomer, 185 kDa Naja kaouthia protein
|
| Buffer: |
10 mM Tris 5 mM MgCl2 10 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2007 Dec 19
|
Insights into complement convertase formation based on the structure of the factor B-cobra venom factor complex
The EMBO Journal 28(16):2469-2478 (2009)
Janssen B, Gomes L, Koning R, Svergun D, Koster A, Fritzinger D, Vogel C, Gros P
|
| RgGuinier |
4.6 |
nm |
| Dmax |
15.0 |
nm |
| VolumePorod |
384 |
nm3 |
|
|
UniProt ID: P17813 (27-256) Endoglin
|
|
|

|
| Sample: |
Endoglin tetramer, 243 kDa Homo sapiens protein
|
| Buffer: |
10 mM Tris–HCl, 50 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2008 Oct 17
|
Structural and functional characterization of soluble endoglin receptor
Biochemical and Biophysical Research Communications 383(4):386-391 (2009)
Le B, Franke D, Svergun D, Han T, Hwang H, Kim K
|
| RgGuinier |
7.6 |
nm |
| Dmax |
26.0 |
nm |
| VolumePorod |
434 |
nm3 |
|
|
UniProt ID: P17813 (27-256) Endoglin
|
|
|

|
| Sample: |
Endoglin dimer, 121 kDa Homo sapiens protein
|
| Buffer: |
10 mM Tris–HCl, 50 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2008 Oct 17
|
Structural and functional characterization of soluble endoglin receptor
Biochemical and Biophysical Research Communications 383(4):386-391 (2009)
Le B, Franke D, Svergun D, Han T, Hwang H, Kim K
|
| RgGuinier |
4.7 |
nm |
| Dmax |
17.0 |
nm |
| VolumePorod |
173 |
nm3 |
|
|
UniProt ID: P50542 (268-602) Peroxisomal targeting signal 1 receptor (C -terminal)
UniProt ID: O75381 (16-80) Peroxisomal membrane protein PEX14 (N-terminal)
UniProt ID: B4E0T2 (22-143) PTS1-BP
|
|
|

|
| Sample: |
Peroxisomal targeting signal 1 receptor (C -terminal) monomer, 48 kDa Homo sapiens protein
Peroxisomal membrane protein PEX14 (N-terminal) monomer, 7 kDa Homo sapiens protein
PTS1-BP monomer, 14 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES-KOH (pH 7.5), 100mM KCl, and 20mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2005 Oct 3
|
Solution structure of human Pex5.Pex14.PTS1 protein complexes obtained by small angle X-ray scattering.
J Biol Chem 284(37):25334-42 (2009)
Shiozawa K, Konarev PV, Neufeld C, Wilmanns M, Svergun DI
|
| RgGuinier |
2.9 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
110 |
nm3 |
|
|
UniProt ID: P50542 (1-639) Peroxisomal targeting signal 1 receptor
|
|
|

|
| Sample: |
Peroxisomal targeting signal 1 receptor monomer, 71 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES-KOH (pH 7.5), 100mM KCl, and 20mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2006 Feb 24
|
Solution structure of human Pex5.Pex14.PTS1 protein complexes obtained by small angle X-ray scattering.
J Biol Chem 284(37):25334-42 (2009)
Shiozawa K, Konarev PV, Neufeld C, Wilmanns M, Svergun DI
|
| RgGuinier |
5.0 |
nm |
| Dmax |
20.0 |
nm |
| VolumePorod |
181 |
nm3 |
|
|
UniProt ID: P50542 (None-None) Peroxisomal targeting signal 1 receptor
UniProt ID: O75381 (16-80) Peroxisomal membrane protein PEX14
UniProt ID: B4E0T2 (22-143) PTS1-BP
|
|
|
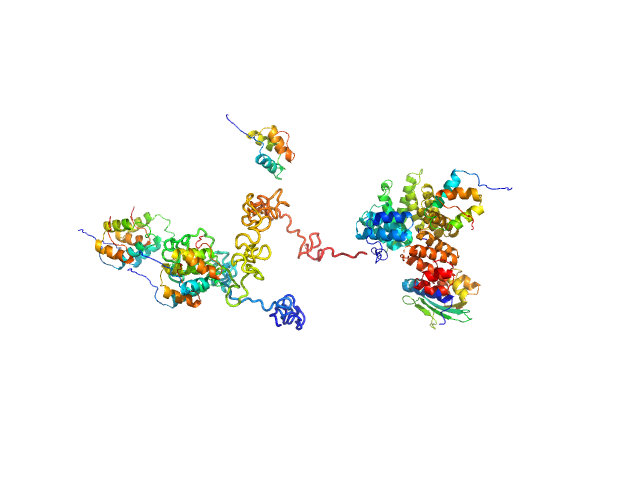
|
| Sample: |
Peroxisomal targeting signal 1 receptor monomer, 71 kDa Homo sapiens protein
Peroxisomal membrane protein PEX14 monomer, 7 kDa Homo sapiens protein
PTS1-BP monomer, 13 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES-KOH (pH 7.5), 100mM KCl, and 20mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2006 Mar 18
|
Solution structure of human Pex5.Pex14.PTS1 protein complexes obtained by small angle X-ray scattering.
J Biol Chem 284(37):25334-42 (2009)
Shiozawa K, Konarev PV, Neufeld C, Wilmanns M, Svergun DI
|
| RgGuinier |
6.0 |
nm |
| Dmax |
20.0 |
nm |
| VolumePorod |
267 |
nm3 |
|
|
UniProt ID: Q7KZI7 (None-None) Serine/threonine-protein kinase MARK2
|
|
|
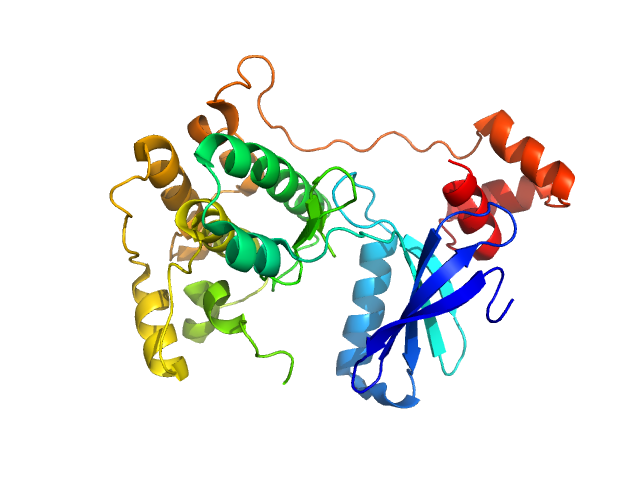
|
| Sample: |
Serine/threonine-protein kinase MARK2 monomer, 36 kDa Homo sapiens protein
|
| Buffer: |
0.1 M Bis-Tris, 0.2 M ammonium citrate, 1mM DTT, pH: 6.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2006 May 16
|
Structural Variations in the Catalytic and Ubiquitin-associated Domains of Microtubule-associated Protein/Microtubule Affinity Regulating Kinase (MARK) 1 and MARK2
Journal of Biological Chemistry 281(37):27586-27599 (2006)
Marx A, Nugoor C, Müller J, Panneerselvam S, Timm T, Bilang M, Mylonas E, Svergun D, Mandelkow E, Mandelkow E
|
| RgGuinier |
2.4 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
61 |
nm3 |
|
|
UniProt ID: Q7KZI7 (None-None) Serine/threonine-protein kinase MARK2
|
|
|
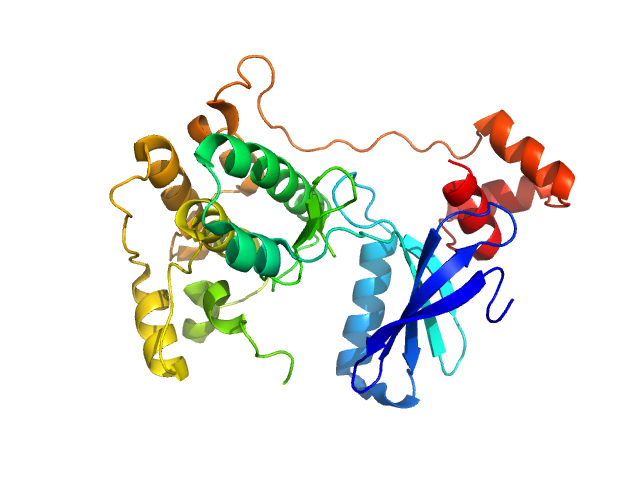
|
| Sample: |
Serine/threonine-protein kinase MARK2 monomer, 36 kDa Homo sapiens protein
|
| Buffer: |
0.1 M Bis-Tris, 0.2 M ammonium citrate, 1mM DTT, pH: 6.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2006 May 16
|
Structural Variations in the Catalytic and Ubiquitin-associated Domains of Microtubule-associated Protein/Microtubule Affinity Regulating Kinase (MARK) 1 and MARK2
Journal of Biological Chemistry 281(37):27586-27599 (2006)
Marx A, Nugoor C, Müller J, Panneerselvam S, Timm T, Bilang M, Mylonas E, Svergun D, Mandelkow E, Mandelkow E
|
| RgGuinier |
2.3 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
62 |
nm3 |
|
|