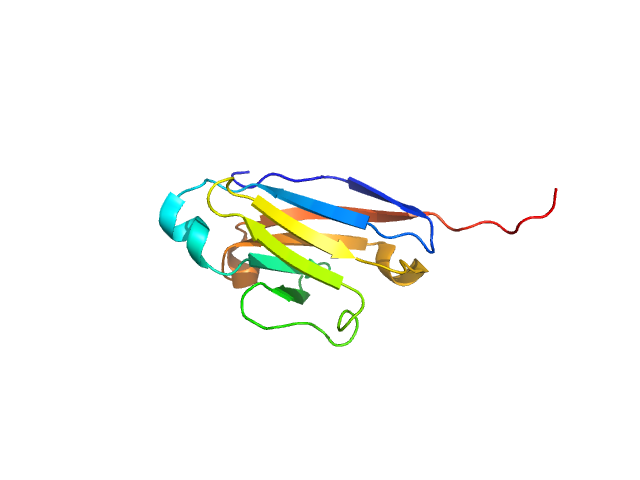
UniProt ID: Q9UGN4 (19-123) CMRF35-like molecule 8
|
|
|
|
| Sample: |
CMRF35-like molecule 8 monomer, 12 kDa Homo sapiens protein
|
| Buffer: |
MES buffer with 3mM DTT, pH: 5.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2006 Apr 6
|
Molecular analysis and solution structure from small-angle X-ray scattering of the human natural killer inhibitory receptor IRp60 (CD300a)
International Journal of Biological Macromolecules 40(3):193-200 (2007)
Dimasi N, Roessle M, Moran O, Candiano G, Svergun D, Biassoni R
|
| RgGuinier |
2.0 |
nm |
| Dmax |
7.0 |
nm |
| VolumePorod |
22 |
nm3 |
|
|
UniProt ID: P39900 (None-None) Macrophage metalloelastase
|
|
|
|
| Sample: |
Macrophage metalloelastase monomer, 54 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris10 mM CaCl2, 0.3 M NaCl, 0.2 M AHA, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2007 Oct 2
|
Evidence of Reciprocal Reorientation of the Catalytic and Hemopexin-Like Domains of Full-Length MMP-12
Journal of the American Chemical Society 130(22):7011-7021 (2008)
Bertini I, Calderone V, Fragai M, Jaiswal R, Luchinat C, Melikian M, Mylonas E, Svergun D
|
| RgGuinier |
3.2 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
58 |
nm3 |
|
|
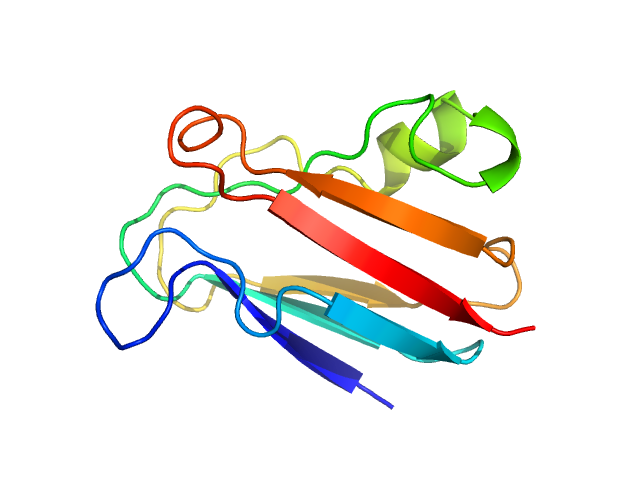
UniProt ID: Q51883 (None-None) Plastocyanin
|
|
|
|
| Sample: |
Plastocyanin monomer, 11 kDa Phormidium laminosum protein
|
| Buffer: |
pure water, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2009 May 7
|
Metal-Mediated Self-Assembly of a β-Sandwich Protein
Chemistry - A European Journal 15(46):12672-12680 (2009)
Crowley P, Matias P, Khan A, Roessle M, Svergun D
|
| RgGuinier |
1.4 |
nm |
| Dmax |
4.5 |
nm |
| VolumePorod |
15 |
nm3 |
|
|
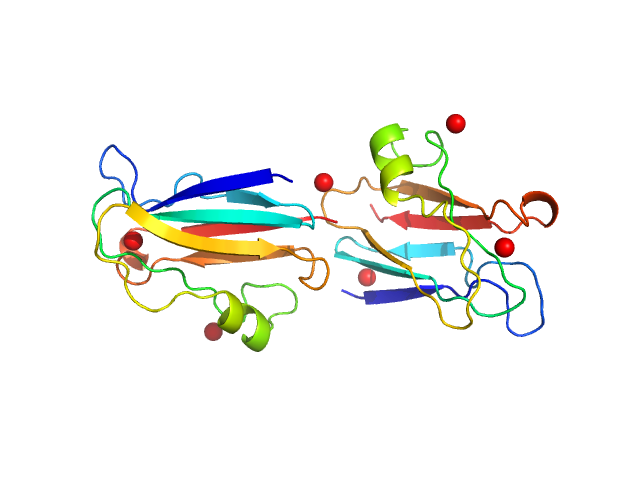
UniProt ID: Q51883 (None-None) Plastocyanin
|
|
|
|
| Sample: |
Plastocyanin dimer, 23 kDa Phormidium laminosum protein
|
| Buffer: |
pure water, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2009 May 7
|
Metal-Mediated Self-Assembly of a β-Sandwich Protein
Chemistry - A European Journal 15(46):12672-12680 (2009)
Crowley P, Matias P, Khan A, Roessle M, Svergun D
|
| RgGuinier |
2.3 |
nm |
| Dmax |
8.5 |
nm |
| VolumePorod |
28 |
nm3 |
|
|
UniProt ID: P62136 (7-330) Serine/threonine-protein phosphatase PP1-alpha catalytic subunit
UniProt ID: Q9UQK1 (70-264) Protein phosphatase 1 regulatory subunit 3C
|
|
|

|
| Sample: |
Serine/threonine-protein phosphatase PP1-alpha catalytic subunit monomer, 37 kDa Homo sapiens protein
Protein phosphatase 1 regulatory subunit 3C monomer, 23 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris pH 8.0, 0.5 M NaCl, 10% glycerol, 1 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2021 Apr 7
|
Molecular architecture of the glycogen- committed PP1/PTG holoenzyme.
Nat Commun 13(1):6199 (2022)
Semrau MS, Giachin G, Covaceuszach S, Cassetta A, Demitri N, Storici P, Lolli G
|
| RgGuinier |
3.3 |
nm |
| Dmax |
11.9 |
nm |
| VolumePorod |
87 |
nm3 |
|
|
UniProt ID: P62136 (7-330) Serine/threonine-protein phosphatase PP1-alpha catalytic subunit
UniProt ID: Q9UQK1 (70-264) Protein phosphatase 1 regulatory subunit 3C
|
|
|

|
| Sample: |
Serine/threonine-protein phosphatase PP1-alpha catalytic subunit monomer, 37 kDa Homo sapiens protein
Protein phosphatase 1 regulatory subunit 3C monomer, 23 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris pH 8.0, 0.5 M NaCl, 10% glycerol, 1 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2021 Apr 7
|
Molecular architecture of the glycogen- committed PP1/PTG holoenzyme.
Nat Commun 13(1):6199 (2022)
Semrau MS, Giachin G, Covaceuszach S, Cassetta A, Demitri N, Storici P, Lolli G
|
| RgGuinier |
3.6 |
nm |
| Dmax |
12.3 |
nm |
| VolumePorod |
89 |
nm3 |
|
|
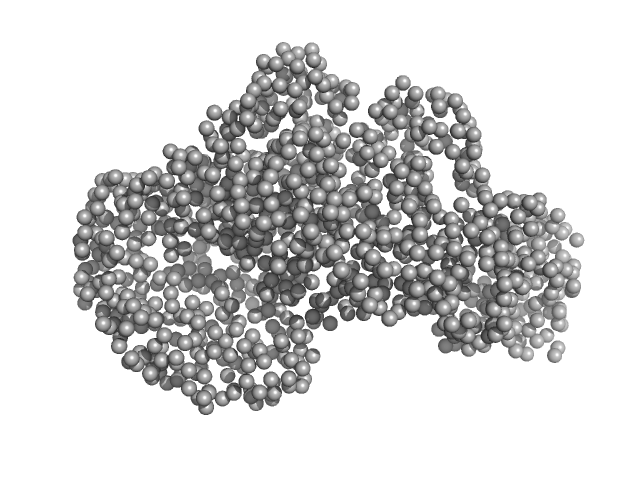
UniProt ID: Q74DF5 (1-473) Piwi domain protein
UniProt ID: Q74DF6 (1-587) Sir2 superfamily protein
|
|
|
|
| Sample: |
Piwi domain protein monomer, 53 kDa Geobacter sulfurreducens (strain … protein
Sir2 superfamily protein monomer, 68 kDa Geobacter sulfurreducens (strain … protein
|
| Buffer: |
20 mM TrisHCl, pH 7.5, 5 mM MgCl2, 150 mM NaCl and 2 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Sep 27
|
Short prokaryotic Argonautes provide defence against incoming mobile genetic elements through NAD+ depletion
Nature Microbiology (2022)
Zaremba M, Dakineviciene D, Golovinas E, Zagorskaitė E, Stankunas E, Lopatina A, Sorek R, Manakova E, Ruksenaite A, Silanskas A, Asmontas S, Grybauskas A, Tylenyte U, Jurgelaitis E, Grigaitis R, Timinskas K, Venclovas Č, Siksnys V
|
| RgGuinier |
3.6 |
nm |
| Dmax |
11.4 |
nm |
| VolumePorod |
185 |
nm3 |
|
|
UniProt ID: Q79PF6 (1-284) Circadian clock protein KaiA
UniProt ID: Q79PF5 (1-102) Circadian clock protein KaiB
UniProt ID: Q79PF4 (1-519) Circadian clock protein kinase KaiC (S431D mutant)
|
|
|

|
| Sample: |
Circadian clock protein KaiA dodecamer, 393 kDa Synechococcus elongatus (strain … protein
Circadian clock protein KaiB hexamer, 71 kDa Synechococcus elongatus (strain … protein
Circadian clock protein kinase KaiC (S431D mutant) hexamer, 357 kDa Synechococcus elongatus (strain … protein
|
| Buffer: |
50 mM sodium phosphate buffer, 150 mM NaCl, 5 mM MgCl2, 0.5 mM EDTA, 1 mM ATP, 1 mM DTT, 50 mM arginine, 50 mM glutamine, pH: 7.8 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2017 Dec 8
|
Overall structure of fully assembled cyanobacterial KaiABC circadian clock complex by an integrated experimental-computational approach.
Commun Biol 5(1):184 (2022)
Yunoki Y, Matsumoto A, Morishima K, Martel A, Porcar L, Sato N, Yogo R, Tominaga T, Inoue R, Yagi-Utsumi M, Okuda A, Shimizu M, Urade R, Terauchi K, Kono H, Yagi H, Kato K, Sugiyama M
|
| RgGuinier |
7.0 |
nm |
| Dmax |
24.5 |
nm |
| VolumePorod |
1650 |
nm3 |
|
|
UniProt ID: Q79PF5 (1-102) 75% deuterated Circadian clock protein KaiB
UniProt ID: Q79PF6 (1-284) Circadian clock protein KaiA
UniProt ID: Q79PF4 (1-519) 75% deuterated Circadian clock protein kinase KaiC (S431D mutant)
|
|
|

|
| Sample: |
75% deuterated Circadian clock protein KaiB hexamer, 71 kDa Synechococcus elongatus (strain … protein
Circadian clock protein KaiA dodecamer, 393 kDa Synechococcus elongatus (strain … protein
75% deuterated Circadian clock protein kinase KaiC (S431D mutant) hexamer, 357 kDa Synechococcus elongatus (strain … protein
|
| Buffer: |
50 mM sodium phosphate buffer, 150 mm NaCl, 5 mM MgCl2, 0.5 mM EDTA, 1 mM ATP, 1 mM DTT, 50 mM arginine, 50 mM glutamine, in 100% D2O, pH: 7.8 |
| Experiment: |
SANS
data collected at D22, Institut Laue-Langevin (ILL) on 2018 Sep 19
|
Overall structure of fully assembled cyanobacterial KaiABC circadian clock complex by an integrated experimental-computational approach.
Commun Biol 5(1):184 (2022)
Yunoki Y, Matsumoto A, Morishima K, Martel A, Porcar L, Sato N, Yogo R, Tominaga T, Inoue R, Yagi-Utsumi M, Okuda A, Shimizu M, Urade R, Terauchi K, Kono H, Yagi H, Kato K, Sugiyama M
|
| RgGuinier |
7.8 |
nm |
| Dmax |
25.6 |
nm |
| VolumePorod |
1620 |
nm3 |
|
|
UniProt ID: P69905 (3-142) Hemoglobin subunit alpha
UniProt ID: P68871 (3-147) Hemoglobin subunit beta
|
|
|
|
| Sample: |
Hemoglobin subunit alpha monomer, 15 kDa Homo sapiens protein
Hemoglobin subunit beta monomer, 16 kDa Homo sapiens protein
|
| Buffer: |
Ringer's lactate solution, pH: 6.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2006 Feb 19
|
Solution Structure of Poly(ethylene) Glycol-Conjugated Hemoglobin Revealed by Small-Angle X-Ray Scattering: Implications for a New Oxygen Therapeutic
Biophysical Journal 94(1):173-181 (2008)
Svergun D, Ekström F, Vandegriff K, Malavalli A, Baker D, Nilsson C, Winslow R
|
| RgGuinier |
2.4 |
nm |
| Dmax |
13.0 |
nm |
|
|