UniProt ID: Q61330 (29-608) Contactin-2
|
|
|
|
| Sample: |
Contactin-2 dimer, 157 kDa Mus musculus protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Dec 7
|
Contactin 2 homophilic adhesion structure and conformational plasticity
Structure (2023)
Chataigner L, Thärichen L, Beugelink J, Granneman J, Mokiem N, Snijder J, Förster F, Janssen B
|
| RgGuinier |
5.8 |
nm |
| Dmax |
32.0 |
nm |
| VolumePorod |
412 |
nm3 |
|
|
UniProt ID: Q61330 (29-1004) Contactin-2
|
|
|
|
| Sample: |
Contactin-2 trimer, 389 kDa Mus musculus protein
|
| Buffer: |
25 mM HEPES, 500 mM NaCl, 500 mM imidazole, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Dec 17
|
Contactin 2 homophilic adhesion structure and conformational plasticity
Structure (2023)
Chataigner L, Thärichen L, Beugelink J, Granneman J, Mokiem N, Snijder J, Förster F, Janssen B
|
| RgGuinier |
9.3 |
nm |
| Dmax |
45.0 |
nm |
| VolumePorod |
780 |
nm3 |
|
|
UniProt ID: Q61330 (29-1004) Contactin-2
|
|
|
|
| Sample: |
Contactin-2 trimer, 389 kDa Mus musculus protein
|
| Buffer: |
25 mM HEPES, 500 mM NaCl, 500 mM imidazole, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Dec 17
|
Contactin 2 homophilic adhesion structure and conformational plasticity
Structure (2023)
Chataigner L, Thärichen L, Beugelink J, Granneman J, Mokiem N, Snijder J, Förster F, Janssen B
|
| RgGuinier |
10.3 |
nm |
| Dmax |
60.0 |
nm |
| VolumePorod |
1290 |
nm3 |
|
|
UniProt ID: Q61330 (29-1004) Contactin-2
|
|
|
|
| Sample: |
Contactin-2 trimer, 389 kDa Mus musculus protein
|
| Buffer: |
25 mM HEPES, 500 mM NaCl, 500 mM imidazole, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Dec 17
|
Contactin 2 homophilic adhesion structure and conformational plasticity
Structure (2023)
Chataigner L, Thärichen L, Beugelink J, Granneman J, Mokiem N, Snijder J, Förster F, Janssen B
|
| RgGuinier |
11.2 |
nm |
| Dmax |
60.0 |
nm |
| VolumePorod |
1540 |
nm3 |
|
|
UniProt ID: Q61330 (29-1004) Contactin-2
|
|
|
|
| Sample: |
Contactin-2 trimer, 389 kDa Mus musculus protein
|
| Buffer: |
25 mM HEPES, 500 mM NaCl, 500 mM imidazole, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Dec 17
|
Contactin 2 homophilic adhesion structure and conformational plasticity
Structure (2023)
Chataigner L, Thärichen L, Beugelink J, Granneman J, Mokiem N, Snijder J, Förster F, Janssen B
|
| RgGuinier |
11.0 |
nm |
| Dmax |
70.0 |
nm |
| VolumePorod |
1900 |
nm3 |
|
|
UniProt ID: Q61330 (29-1004) Contactin-2
|
|
|
|
| Sample: |
Contactin-2 trimer, 389 kDa Mus musculus protein
|
| Buffer: |
25 mM HEPES, 500 mM NaCl, 500 mM imidazole, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Dec 17
|
Contactin 2 homophilic adhesion structure and conformational plasticity
Structure (2023)
Chataigner L, Thärichen L, Beugelink J, Granneman J, Mokiem N, Snijder J, Förster F, Janssen B
|
| RgGuinier |
11.3 |
nm |
| Dmax |
66.0 |
nm |
| VolumePorod |
1820 |
nm3 |
|
|
UniProt ID: Q61330 (29-1004) Contactin-2
|
|
|
|
| Sample: |
Contactin-2 trimer, 389 kDa Mus musculus protein
|
| Buffer: |
25 mM HEPES, 500 mM NaCl, 500 mM imidazole, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Dec 17
|
Contactin 2 homophilic adhesion structure and conformational plasticity
Structure (2023)
Chataigner L, Thärichen L, Beugelink J, Granneman J, Mokiem N, Snijder J, Förster F, Janssen B
|
| RgGuinier |
12.5 |
nm |
| Dmax |
65.0 |
nm |
| VolumePorod |
2044 |
nm3 |
|
|
UniProt ID: Q61330 (29-1004) Contactin-2
|
|
|
|
| Sample: |
Contactin-2 trimer, 389 kDa Mus musculus protein
|
| Buffer: |
25 mM HEPES, 500 mM NaCl, 500 mM imidazole, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Dec 17
|
Contactin 2 homophilic adhesion structure and conformational plasticity
Structure (2023)
Chataigner L, Thärichen L, Beugelink J, Granneman J, Mokiem N, Snijder J, Förster F, Janssen B
|
| RgGuinier |
14.2 |
nm |
| Dmax |
65.0 |
nm |
| VolumePorod |
2190 |
nm3 |
|
|
UniProt ID: A0A0J0ZLX8 (1-446) L-lysine 6-monooxygenase (NADPH-requiring)
|
|
|
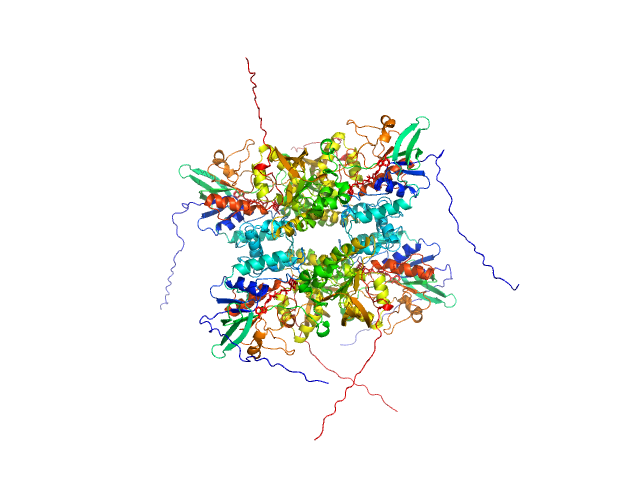
|
| Sample: |
L-lysine 6-monooxygenase (NADPH-requiring) tetramer, 214 kDa Acinetobacter baumannii MRSN … protein
|
| Buffer: |
25 mM HEPES pH 7.5, 150 mM NaCl, 1 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2022 Apr 14
|
Kinetic and Structural Characterization of a Flavin-Dependent Putrescine N-Hydroxylase from Acinetobacter baumannii.
Biochemistry (2022)
Lyons NS, Bogner AN, Tanner JJ, Sobrado P
|
| RgGuinier |
4.1 |
nm |
| Dmax |
15.0 |
nm |
| VolumePorod |
394 |
nm3 |
|
|
UniProt ID: A0A0J0ZLX8 (1-446) L-lysine 6-monooxygenase (NADPH-requiring)
|
|
|
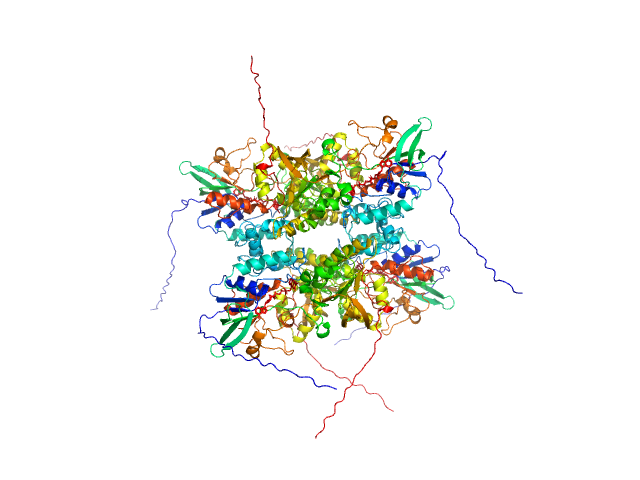
|
| Sample: |
L-lysine 6-monooxygenase (NADPH-requiring) tetramer, 214 kDa Acinetobacter baumannii MRSN … protein
|
| Buffer: |
25 mM HEPES pH 7.5, 150 mM NaCl, 1 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2022 Apr 14
|
Kinetic and Structural Characterization of a Flavin-Dependent Putrescine N-Hydroxylase from Acinetobacter baumannii.
Biochemistry (2022)
Lyons NS, Bogner AN, Tanner JJ, Sobrado P
|
| RgGuinier |
4.1 |
nm |
| Dmax |
15.0 |
nm |
| VolumePorod |
390 |
nm3 |
|
|