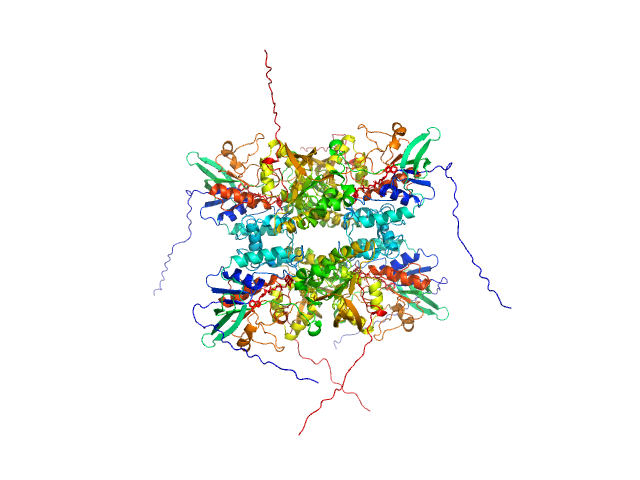
UniProt ID: A0A0J0ZLX8 (1-446) L-lysine 6-monooxygenase (NADPH-requiring)
|
|
|
|
| Sample: |
L-lysine 6-monooxygenase (NADPH-requiring) tetramer, 214 kDa Acinetobacter baumannii MRSN … protein
|
| Buffer: |
25 mM HEPES pH 7.5, 150 mM NaCl, 1 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2022 Apr 14
|
Kinetic and Structural Characterization of a Flavin-Dependent Putrescine N-Hydroxylase from Acinetobacter baumannii.
Biochemistry (2022)
Lyons NS, Bogner AN, Tanner JJ, Sobrado P
|
| RgGuinier |
4.2 |
nm |
| Dmax |
15.0 |
nm |
| VolumePorod |
390 |
nm3 |
|
|
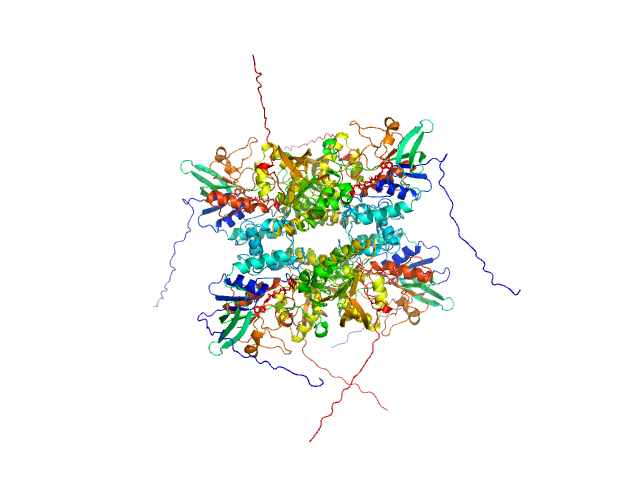
UniProt ID: A0A0J0ZLX8 (1-446) L-lysine 6-monooxygenase (NADPH-requiring)
|
|
|
|
| Sample: |
L-lysine 6-monooxygenase (NADPH-requiring) tetramer, 214 kDa Acinetobacter baumannii MRSN … protein
|
| Buffer: |
25 mM HEPES pH 7.5, 150 mM NaCl, 1 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2022 Apr 14
|
Kinetic and Structural Characterization of a Flavin-Dependent Putrescine N-Hydroxylase from Acinetobacter baumannii.
Biochemistry (2022)
Lyons NS, Bogner AN, Tanner JJ, Sobrado P
|
| RgGuinier |
4.2 |
nm |
| Dmax |
15.4 |
nm |
| VolumePorod |
390 |
nm3 |
|
|
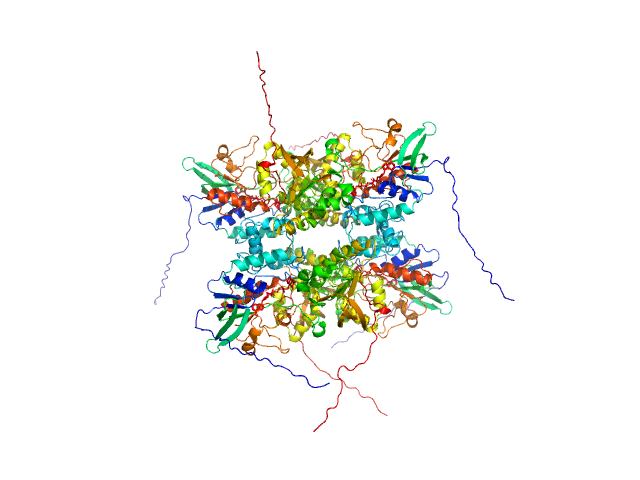
UniProt ID: A0A0J0ZLX8 (1-446) L-lysine 6-monooxygenase (NADPH-requiring)
|
|
|
|
| Sample: |
L-lysine 6-monooxygenase (NADPH-requiring) tetramer, 214 kDa Acinetobacter baumannii MRSN … protein
|
| Buffer: |
25 mM HEPES pH 7.5, 150 mM NaCl, 1 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2022 Apr 14
|
Kinetic and Structural Characterization of a Flavin-Dependent Putrescine N-Hydroxylase from Acinetobacter baumannii.
Biochemistry (2022)
Lyons NS, Bogner AN, Tanner JJ, Sobrado P
|
| RgGuinier |
4.2 |
nm |
| Dmax |
15.5 |
nm |
| VolumePorod |
390 |
nm3 |
|
|
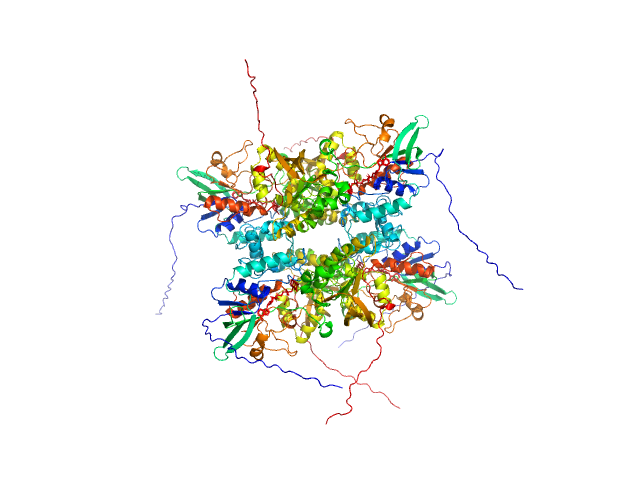
UniProt ID: A0A0J0ZLX8 (1-446) L-lysine 6-monooxygenase (NADPH-requiring)
|
|
|
|
| Sample: |
L-lysine 6-monooxygenase (NADPH-requiring) tetramer, 214 kDa Acinetobacter baumannii MRSN … protein
|
| Buffer: |
25 mM HEPES pH 7.5, 150 mM NaCl, 1 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2022 Apr 14
|
Kinetic and Structural Characterization of a Flavin-Dependent Putrescine N-Hydroxylase from Acinetobacter baumannii.
Biochemistry (2022)
Lyons NS, Bogner AN, Tanner JJ, Sobrado P
|
| RgGuinier |
4.2 |
nm |
| Dmax |
15.0 |
nm |
| VolumePorod |
396 |
nm3 |
|
|
UniProt ID: Q9NXU5 (29-204) ADP-ribosylation factor-like protein 15
|
|
|

|
| Sample: |
ADP-ribosylation factor-like protein 15 monomer, 22 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 100 mM NaCl, 1 mM PMSF, 10% glycerol, 1 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpace, CSIR - Institute of Microbial Technology (IMTech) on 2018 Jul 10
|
ARL15, a GTPase implicated in rheumatoid arthritis, potentially repositions its truncated N-terminus as a function of guanine nucleotide binding
International Journal of Biological Macromolecules 254:127898 (2024)
Saini M, Upadhyay N, Dhiman K, Manjhi S, Kattuparambil A, Ghoshal A, Arya R, Dey S, Sharma A, Aduri R, Thelma B, Ashish F, Kundu S
|
| RgGuinier |
1.8 |
nm |
| Dmax |
6.1 |
nm |
| VolumePorod |
33 |
nm3 |
|
|
UniProt ID: Q9NXU5 (29-204) ADP-ribosylation factor-like protein 15
|
|
|
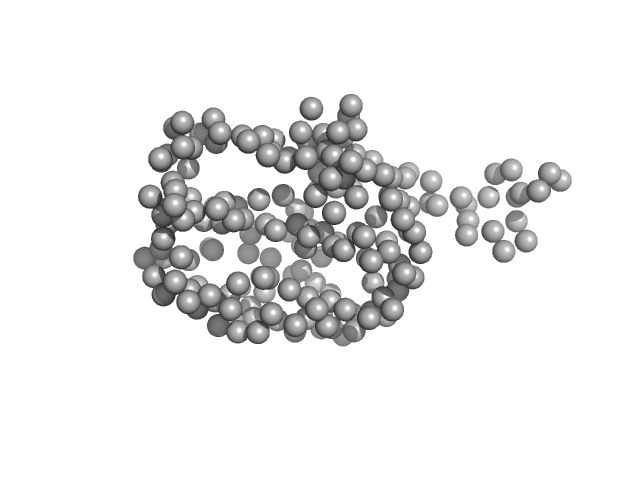
|
| Sample: |
ADP-ribosylation factor-like protein 15 monomer, 22 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 100 mM NaCl, 1 mM PMSF, 10% glycerol, 1 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpace, CSIR - Institute of Microbial Technology (IMTech) on 2018 Jul 10
|
ARL15, a GTPase implicated in rheumatoid arthritis, potentially repositions its truncated N-terminus as a function of guanine nucleotide binding
International Journal of Biological Macromolecules 254:127898 (2024)
Saini M, Upadhyay N, Dhiman K, Manjhi S, Kattuparambil A, Ghoshal A, Arya R, Dey S, Sharma A, Aduri R, Thelma B, Ashish F, Kundu S
|
| RgGuinier |
2.0 |
nm |
| Dmax |
7.6 |
nm |
| VolumePorod |
38 |
nm3 |
|
|
UniProt ID: Q9NXU5 (29-204) ADP-ribosylation factor-like protein 15
|
|
|

|
| Sample: |
ADP-ribosylation factor-like protein 15 monomer, 22 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 100 mM NaCl, 1 mM PMSF, 10% glycerol, 1 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpace, CSIR - Institute of Microbial Technology (IMTech) on 2018 Jul 10
|
ARL15, a GTPase implicated in rheumatoid arthritis, potentially repositions its truncated N-terminus as a function of guanine nucleotide binding
International Journal of Biological Macromolecules 254:127898 (2024)
Saini M, Upadhyay N, Dhiman K, Manjhi S, Kattuparambil A, Ghoshal A, Arya R, Dey S, Sharma A, Aduri R, Thelma B, Ashish F, Kundu S
|
| RgGuinier |
2.0 |
nm |
| Dmax |
7.7 |
nm |
| VolumePorod |
38 |
nm3 |
|
|
UniProt ID: I6X8D2 (1-1733) Polyketide synthase Pks13
|
|
|

|
| Sample: |
Polyketide synthase Pks13 monomer, 188 kDa Mycobacterium tuberculosis (strain … protein
|
| Buffer: |
50 mM Tris-HCl, 50 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2013 Mar 1
|
Solution structure of the type I polyketide synthase Pks13 from Mycobacterium tuberculosis.
BMC Biol 20(1):147 (2022)
Bon C, Cabantous S, Julien S, Guillet V, Chalut C, Rima J, Brison Y, Malaga W, Sanchez-Dafun A, Gavalda S, Quémard A, Marcoux J, Waldo GS, Guilhot C, Mourey L
|
| RgGuinier |
7.3 |
nm |
| Dmax |
24.8 |
nm |
| VolumePorod |
480 |
nm3 |
|
|
UniProt ID: I6X8D2 (1-1733) Polyketide synthase Pks13
|
|
|
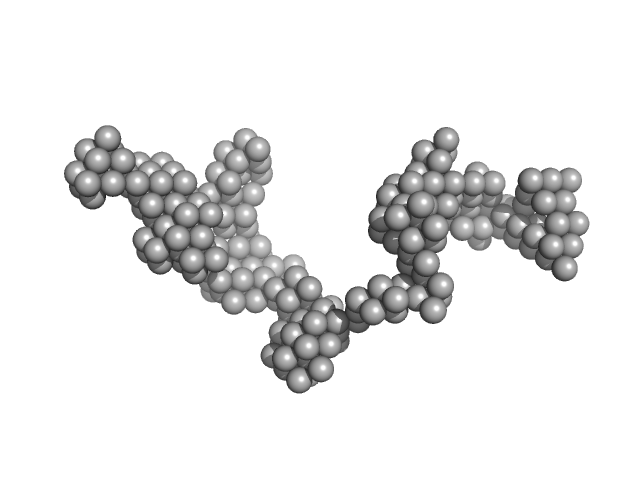
|
| Sample: |
Polyketide synthase Pks13 dimer, 376 kDa Mycobacterium tuberculosis (strain … protein
|
| Buffer: |
50 mM Tris-HCl, 50 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2011 Nov 23
|
Solution structure of the type I polyketide synthase Pks13 from Mycobacterium tuberculosis.
BMC Biol 20(1):147 (2022)
Bon C, Cabantous S, Julien S, Guillet V, Chalut C, Rima J, Brison Y, Malaga W, Sanchez-Dafun A, Gavalda S, Quémard A, Marcoux J, Waldo GS, Guilhot C, Mourey L
|
| RgGuinier |
10.3 |
nm |
| Dmax |
36.0 |
nm |
| VolumePorod |
2300 |
nm3 |
|
|
UniProt ID: I6X8D2 (1437-1725) Polyketide synthase Pks13
|
|
|

|
| Sample: |
Polyketide synthase Pks13 monomer, 33 kDa Mycobacterium tuberculosis (strain … protein
|
| Buffer: |
50 mM Tris-HCl, 50 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2006 Nov 1
|
Solution structure of the type I polyketide synthase Pks13 from Mycobacterium tuberculosis.
BMC Biol 20(1):147 (2022)
Bon C, Cabantous S, Julien S, Guillet V, Chalut C, Rima J, Brison Y, Malaga W, Sanchez-Dafun A, Gavalda S, Quémard A, Marcoux J, Waldo GS, Guilhot C, Mourey L
|
| RgGuinier |
1.9 |
nm |
| Dmax |
5.5 |
nm |
| VolumePorod |
56 |
nm3 |
|
|