UniProt ID: P02769 (1-607) Albumin
|
|
|
|
| Sample: |
Albumin monomer, 69 kDa Bos taurus protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, and 1% v/v glycerol, pH: 7 |
| Experiment: |
SAXS
data collected at ID7A1 BioSAXS / HP-Bio Beamline, Cornell High Energy Synchrotron Source (CHESS) on 2020 Mar 7
|
Inline small‐angle X‐ray scattering‐coupled chromatography under extreme hydrostatic pressure
Protein Science 31(12) (2022)
Miller R, Cummings C, Huang Q, Ando N, Gillilan R
|
| RgGuinier |
3.0 |
nm |
| Dmax |
9.5 |
nm |
| VolumePorod |
108 |
nm3 |
|
|
UniProt ID: P02769 (1-607) Albumin
|
|
|
|
| Sample: |
Albumin monomer, 69 kDa Bos taurus protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, and 1% v/v glycerol, pH: 7 |
| Experiment: |
SAXS
data collected at ID7A1 BioSAXS / HP-Bio Beamline, Cornell High Energy Synchrotron Source (CHESS) on 2020 Mar 7
|
Inline small‐angle X‐ray scattering‐coupled chromatography under extreme hydrostatic pressure
Protein Science 31(12) (2022)
Miller R, Cummings C, Huang Q, Ando N, Gillilan R
|
| RgGuinier |
2.8 |
nm |
| Dmax |
9.4 |
nm |
| VolumePorod |
104 |
nm3 |
|
|
UniProt ID: P39687 (1-249) Acidic leucine-rich nuclear phosphoprotein 32 family member A (L60A mutant)
|
|
|
|
| Sample: |
Acidic leucine-rich nuclear phosphoprotein 32 family member A (L60A mutant) monomer, 29 kDa Homo sapiens protein
|
| Buffer: |
25 mM BisTRIS, 10 mM NaCl, 5 mM DTT, pH: 6.8 |
| Experiment: |
SAXS
data collected at ID7A1 BioSAXS / HP-Bio Beamline, Cornell High Energy Synchrotron Source (CHESS) on 2020 Mar 7
|
Inline small‐angle X‐ray scattering‐coupled chromatography under extreme hydrostatic pressure
Protein Science 31(12) (2022)
Miller R, Cummings C, Huang Q, Ando N, Gillilan R
|
| RgGuinier |
1.8 |
nm |
| Dmax |
6.0 |
nm |
| VolumePorod |
31 |
nm3 |
|
|
UniProt ID: P39687 (1-249) Acidic leucine-rich nuclear phosphoprotein 32 family member A (L60A mutant)
|
|
|
|
| Sample: |
Acidic leucine-rich nuclear phosphoprotein 32 family member A (L60A mutant) monomer, 29 kDa Homo sapiens protein
|
| Buffer: |
25 mM BisTRIS, 10 mM NaCl, 5 mM DTT, pH: 6.8 |
| Experiment: |
SAXS
data collected at ID7A1 BioSAXS / HP-Bio Beamline, Cornell High Energy Synchrotron Source (CHESS) on 2020 Mar 7
|
Inline small‐angle X‐ray scattering‐coupled chromatography under extreme hydrostatic pressure
Protein Science 31(12) (2022)
Miller R, Cummings C, Huang Q, Ando N, Gillilan R
|
| RgGuinier |
2.0 |
nm |
| Dmax |
7.5 |
nm |
| VolumePorod |
37 |
nm3 |
|
|
UniProt ID: P32566 (80-340) Cell wall assembly regulator SMI1
|
|
|

|
| Sample: |
Cell wall assembly regulator SMI1 monomer, 31 kDa Saccharomyces cerevisiae (strain … protein
|
| Buffer: |
100 mM MES, 300 mM NaCl, pH: 6 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Mar 18
|
The conserved yeast protein Knr4 involved in cell wall integrity is a multi-domain intrinsically disordered protein.
J Mol Biol :168048 (2023)
Batista M, Donker EIM, Bon C, Guillien M, Caisso A, Mourey Funding L, Marie François Funding J, Maveyraud L, Zerbib D
|
| RgGuinier |
2.3 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
62 |
nm3 |
|
|
UniProt ID: P32566 (1-345) Cell wall assembly regulator SMI1
|
|
|

|
| Sample: |
Cell wall assembly regulator SMI1 monomer, 40 kDa Saccharomyces cerevisiae (strain … protein
|
| Buffer: |
100 mM MES, 300 mM NaCl, pH: 6 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Mar 4
|
The conserved yeast protein Knr4 involved in cell wall integrity is a multi-domain intrinsically disordered protein.
J Mol Biol :168048 (2023)
Batista M, Donker EIM, Bon C, Guillien M, Caisso A, Mourey Funding L, Marie François Funding J, Maveyraud L, Zerbib D
|
| RgGuinier |
2.8 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
79 |
nm3 |
|
|
UniProt ID: P32566 (1-505) Cell wall assembly regulator SMI1
|
|
|
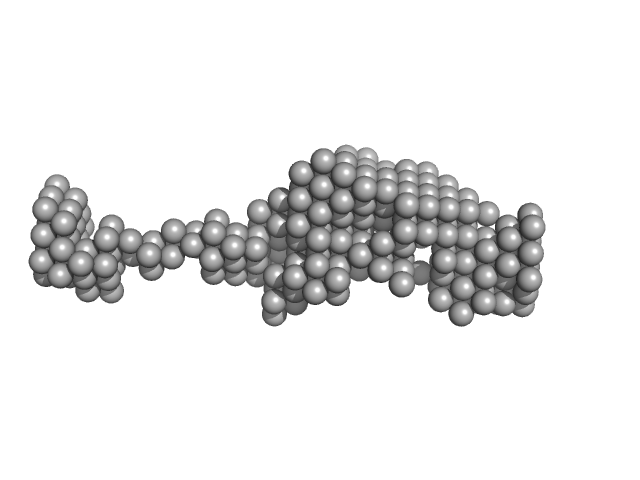
|
| Sample: |
Cell wall assembly regulator SMI1 monomer, 57 kDa Saccharomyces cerevisiae (strain … protein
|
| Buffer: |
100 mM MES, 300 mM NaCl, pH: 6 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Mar 7
|
The conserved yeast protein Knr4 involved in cell wall integrity is a multi-domain intrinsically disordered protein.
J Mol Biol :168048 (2023)
Batista M, Donker EIM, Bon C, Guillien M, Caisso A, Mourey Funding L, Marie François Funding J, Maveyraud L, Zerbib D
|
| RgGuinier |
4.8 |
nm |
| Dmax |
17.0 |
nm |
| VolumePorod |
117 |
nm3 |
|
|
UniProt ID: P61823 (27-150) Ribonuclease pancreatic
|
|
|
|
| Sample: |
Ribonuclease pancreatic monomer, 14 kDa Bos taurus protein
|
| Buffer: |
50 mM HEPES, 150 mM KCl, 3% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Sep 16
|
Small-Angle X-ray Scattering data (benchmarking/consensus): EMBL-P12 SAXS beam line, DESY
Clement Blanchet,
Melissa Graewert,
Cy M Jeffries,
Dmitri Svergun
|
| RgGuinier |
1.5 |
nm |
| Dmax |
4.6 |
nm |
| VolumePorod |
16 |
nm3 |
|
|
UniProt ID: P61823 (27-150) Ribonuclease pancreatic
|
|
|
|
| Sample: |
Ribonuclease pancreatic monomer, 14 kDa Bos taurus protein
|
| Buffer: |
50 mM Tris, 100 mM NaCl, 1% v/v glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Jul 8
|
Small-Angle X-ray Scattering data (benchmarking/consensus): EMBL-P12 SAXS beam line, DESY
Clement Blanchet,
Melissa Graewert,
Cy M Jeffries,
Dmitri Svergun
|
| RgGuinier |
1.5 |
nm |
| Dmax |
4.8 |
nm |
| VolumePorod |
16 |
nm3 |
|
|
UniProt ID: Q00511 (1-302) Urate Oxidase (Uricase) from Aspergillus flavus
|
|
|
|
| Sample: |
Urate Oxidase (Uricase) from Aspergillus flavus tetramer, 137 kDa Aspergillus flavus protein
|
| Buffer: |
100 mM Tris, 150 mM NaCl, 1 % v/v glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Sep 15
|
Small-Angle X-ray Scattering data (benchmarking/consensus): EMBL-P12 SAXS beam line, DESY
Clement Blanchet,
Melissa Graewert,
Cy M Jeffries,
Dmitri Svergun
|
| RgGuinier |
3.2 |
nm |
| Dmax |
8.8 |
nm |
| VolumePorod |
221 |
nm3 |
|
|