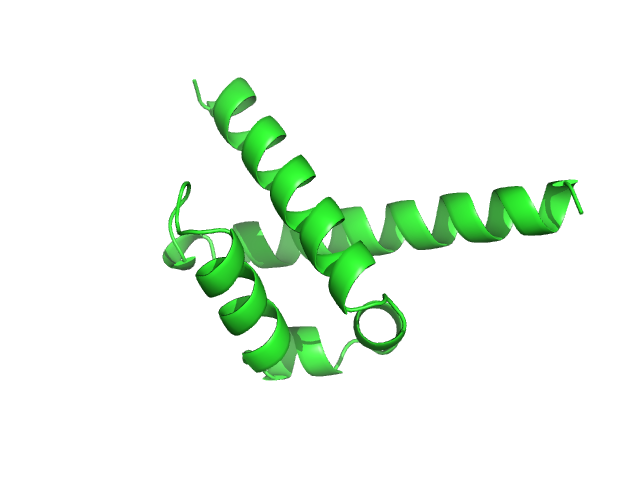
UniProt ID: O75400 (516-592) Pre-mRNA-processing factor 40 homolog A
|
|
|
|
| Sample: |
Pre-mRNA-processing factor 40 homolog A monomer, 9 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, 100 mM NaCl, 2 mM CaCl2, 1 mM TCEP, pH: 7.4 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2021 Sep 23
|
Binding by calmodulin is coupled to transient unfolding of the third FF domain of Prp40A.
Protein Sci 32(4):e4606 (2023)
Díaz Casas A, Cordoba JJ, Ferrer BJ, Balakrishnan S, Wurm JE, Pastrana-Ríos B, Chazin WJ
|
| RgGuinier |
1.6 |
nm |
| Dmax |
7.2 |
nm |
| VolumePorod |
13 |
nm3 |
|
|
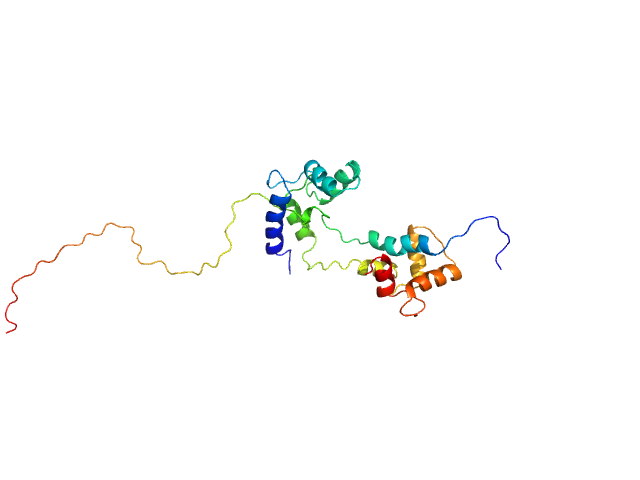
UniProt ID: P0DP23 (1-149) Calmodulin-1
UniProt ID: O75400 (516-592) Pre-mRNA-processing factor 40 homolog A
|
|
|
|
| Sample: |
Calmodulin-1 monomer, 17 kDa Homo sapiens protein
Pre-mRNA-processing factor 40 homolog A monomer, 9 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, 100 mM NaCl, 2 mM CaCl2, 1 mM TCEP, pH: 7.4 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2021 Sep 23
|
Binding by calmodulin is coupled to transient unfolding of the third FF domain of Prp40A.
Protein Sci 32(4):e4606 (2023)
Díaz Casas A, Cordoba JJ, Ferrer BJ, Balakrishnan S, Wurm JE, Pastrana-Ríos B, Chazin WJ
|
| RgGuinier |
2.4 |
nm |
| Dmax |
8.7 |
nm |
| VolumePorod |
29 |
nm3 |
|
|
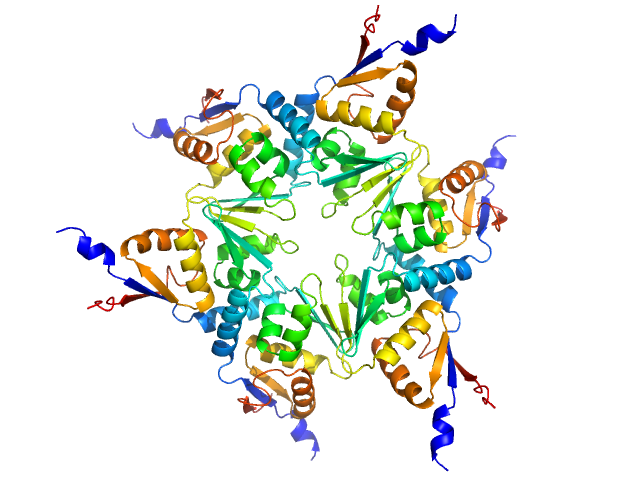
UniProt ID: P42283 (1-120) Longitudinals lacking protein, isoform G
|
|
|
|
| Sample: |
Longitudinals lacking protein, isoform G hexamer, 92 kDa Drosophila melanogaster protein
|
| Buffer: |
20 mM Tris, pH 7.4, 200 mM NaCl, 1 mM DTT, pH: 7.4 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Jul 8
|
BTB domains: A structural view of evolution, multimerization, and protein-protein interactions.
Bioessays :e2200179 (2022)
Bonchuk A, Balagurov K, Georgiev P
|
| RgGuinier |
4.1 |
nm |
| Dmax |
20.0 |
nm |
| VolumePorod |
213 |
nm3 |
|
|
UniProt ID: P42283 (1-120) Longitudinals lacking protein, isoform G
|
|
|
|
| Sample: |
Longitudinals lacking protein, isoform G hexamer, 92 kDa Drosophila melanogaster protein
|
| Buffer: |
20 mM Tris, pH 7.4, 200 mM NaCl, 1 mM DTT, pH: 7.4 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Jul 8
|
BTB domains: A structural view of evolution, multimerization, and protein-protein interactions.
Bioessays :e2200179 (2022)
Bonchuk A, Balagurov K, Georgiev P
|
| RgGuinier |
4.8 |
nm |
| Dmax |
24.0 |
nm |
| VolumePorod |
263 |
nm3 |
|
|
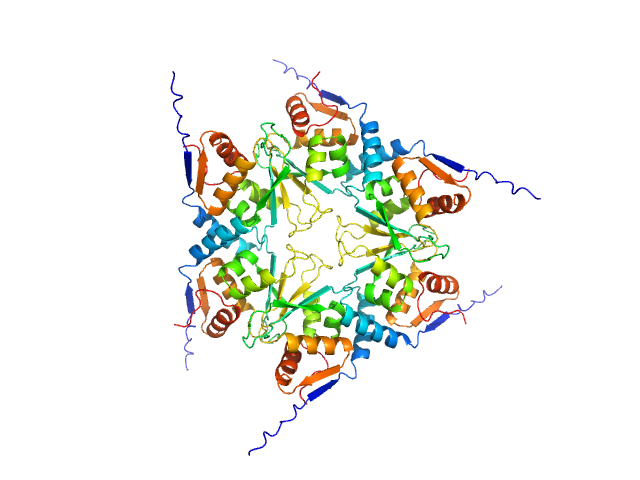
UniProt ID: Q9VSL1 (1-133) Uncharacterized protein, isoform A
|
|
|
|
| Sample: |
Uncharacterized protein, isoform A hexamer, 92 kDa Drosophila melanogaster protein
|
| Buffer: |
20 mM Tris, pH 7.4, 200 mM NaCl, 1 mM DTT, pH: 7.4 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Jul 8
|
BTB domains: A structural view of evolution, multimerization, and protein-protein interactions.
Bioessays :e2200179 (2022)
Bonchuk A, Balagurov K, Georgiev P
|
| RgGuinier |
3.7 |
nm |
| Dmax |
15.0 |
nm |
| VolumePorod |
167 |
nm3 |
|
|
UniProt ID: Q9VSL1 (1-133) Uncharacterized protein, isoform A
|
|
|
|
| Sample: |
Uncharacterized protein, isoform A hexamer, 92 kDa Drosophila melanogaster protein
|
| Buffer: |
20 mM Tris, pH 7.4, 200 mM NaCl, 1 mM DTT, pH: 7.4 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Jul 8
|
BTB domains: A structural view of evolution, multimerization, and protein-protein interactions.
Bioessays :e2200179 (2022)
Bonchuk A, Balagurov K, Georgiev P
|
| RgGuinier |
4.1 |
nm |
| Dmax |
17.0 |
nm |
| VolumePorod |
192 |
nm3 |
|
|
UniProt ID: P24300 (1-380) Xylose isomerase
|
|
|
|
| Sample: |
Xylose isomerase tetramer, 173 kDa Streptomyces rubiginosus protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, 3% v/v glycerol, pH: 7 |
| Experiment: |
SAXS
data collected at ID7A1 BioSAXS / HP-Bio Beamline, Cornell High Energy Synchrotron Source (CHESS) on 2020 Mar 7
|
Inline small‐angle X‐ray scattering‐coupled chromatography under extreme hydrostatic pressure
Protein Science 31(12) (2022)
Miller R, Cummings C, Huang Q, Ando N, Gillilan R
|
| RgGuinier |
3.4 |
nm |
| Dmax |
10.6 |
nm |
| VolumePorod |
253 |
nm3 |
|
|
UniProt ID: P24300 (1-380) Xylose isomerase
|
|
|
|
| Sample: |
Xylose isomerase tetramer, 173 kDa Streptomyces rubiginosus protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, and 3% v/v glycerol, pH: 7 |
| Experiment: |
SAXS
data collected at ID7A1 BioSAXS / HP-Bio Beamline, Cornell High Energy Synchrotron Source (CHESS) on 2020 Mar 7
|
Inline small‐angle X‐ray scattering‐coupled chromatography under extreme hydrostatic pressure
Protein Science 31(12) (2022)
Miller R, Cummings C, Huang Q, Ando N, Gillilan R
|
| RgGuinier |
3.5 |
nm |
| Dmax |
10.6 |
nm |
| VolumePorod |
267 |
nm3 |
|
|
UniProt ID: P02769 (1-607) Albumin
|
|
|
|
| Sample: |
Albumin monomer, 69 kDa Bos taurus protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, and 1% v/v glycerol, pH: 7 |
| Experiment: |
SAXS
data collected at ID7A1 BioSAXS / HP-Bio Beamline, Cornell High Energy Synchrotron Source (CHESS) on 2020 Mar 7
|
Inline small‐angle X‐ray scattering‐coupled chromatography under extreme hydrostatic pressure
Protein Science 31(12) (2022)
Miller R, Cummings C, Huang Q, Ando N, Gillilan R
|
| RgGuinier |
2.7 |
nm |
| Dmax |
8.5 |
nm |
| VolumePorod |
99 |
nm3 |
|
|
UniProt ID: P02769 (1-607) Albumin
|
|
|
|
| Sample: |
Albumin monomer, 69 kDa Bos taurus protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, and 1% v/v glycerol, pH: 7 |
| Experiment: |
SAXS
data collected at ID7A1 BioSAXS / HP-Bio Beamline, Cornell High Energy Synchrotron Source (CHESS) on 2020 Mar 7
|
Inline small‐angle X‐ray scattering‐coupled chromatography under extreme hydrostatic pressure
Protein Science 31(12) (2022)
Miller R, Cummings C, Huang Q, Ando N, Gillilan R
|
| RgGuinier |
2.6 |
nm |
| Dmax |
8.3 |
nm |
| VolumePorod |
94 |
nm3 |
|
|