UniProt ID: Q00511 (1-302) Urate Oxidase (Uricase) from Aspergillus flavus
|
|
|
|
| Sample: |
Urate Oxidase (Uricase) from Aspergillus flavus tetramer, 137 kDa Aspergillus flavus protein
|
| Buffer: |
100 mM Tris, 150 mM NaCl, 1 % v/v glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Jul 8
|
Small-Angle X-ray Scattering data (benchmarking/consensus): EMBL-P12 SAXS beam line, DESY
Clement Blanchet,
Melissa Graewert,
Cy M Jeffries,
Dmitri Svergun
|
| RgGuinier |
3.2 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
214 |
nm3 |
|
|
UniProt ID: P24300 (1-388) Xylose isomerase
|
|
|
|
| Sample: |
Xylose isomerase tetramer, 173 kDa Streptomyces rubiginosus protein
|
| Buffer: |
50 mM Tris, 100 mM NaCl, 1 mM MgCl2, 1 % v/v glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Sep 15
|
Small-Angle X-ray Scattering data (benchmarking/consensus): EMBL-P12 SAXS beam line, DESY
Clement Blanchet,
Melissa Graewert,
Cy M Jeffries,
Dmitri Svergun
|
| RgGuinier |
3.2 |
nm |
| Dmax |
14.1 |
nm |
| VolumePorod |
228 |
nm3 |
|
|
UniProt ID: P24300 (1-388) Xylose isomerase
|
|
|
|
| Sample: |
Xylose isomerase tetramer, 173 kDa Streptomyces rubiginosus protein
|
| Buffer: |
50 mM Tris, 100 mM NaCl, 1 mM MgCl2, 1 % v/v glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Jul 8
|
Small-Angle X-ray Scattering data (benchmarking/consensus): EMBL-P12 SAXS beam line, DESY
Clement Blanchet,
Melissa Graewert,
Cy M Jeffries,
Dmitri Svergun
|
| RgGuinier |
3.3 |
nm |
| Dmax |
14.1 |
nm |
| VolumePorod |
233 |
nm3 |
|
|
UniProt ID: F8W669 (1-190) Endo-1,4-beta-xylanase
|
|
|
|
| Sample: |
Endo-1,4-beta-xylanase monomer, 21 kDa Trichoderma longibrachiatum protein
|
| Buffer: |
50 mM HEPES, 150 mM KCl, 3% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Sep 16
|
Small-Angle X-ray Scattering data (benchmarking/consensus): EMBL-P12 SAXS beam line, DESY
Clement Blanchet,
Melissa Graewert,
Cy M Jeffries,
Dmitri Svergun
|
| RgGuinier |
1.6 |
nm |
| Dmax |
4.8 |
nm |
| VolumePorod |
25 |
nm3 |
|
|
UniProt ID: F8W669 (1-190) Endo-1,4-beta-xylanase
|
|
|
|
| Sample: |
Endo-1,4-beta-xylanase monomer, 21 kDa Trichoderma longibrachiatum protein
|
| Buffer: |
50 mM Tris, 100 mM NaCl, 1% v/v glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Jul 8
|
Small-Angle X-ray Scattering data (benchmarking/consensus): EMBL-P12 SAXS beam line, DESY
Clement Blanchet,
Melissa Graewert,
Cy M Jeffries,
Dmitri Svergun
|
| RgGuinier |
1.6 |
nm |
| Dmax |
4.5 |
nm |
| VolumePorod |
24 |
nm3 |
|
|
UniProt ID: P05067 (672-713) Amyloid-beta precursor protein
|
|
|
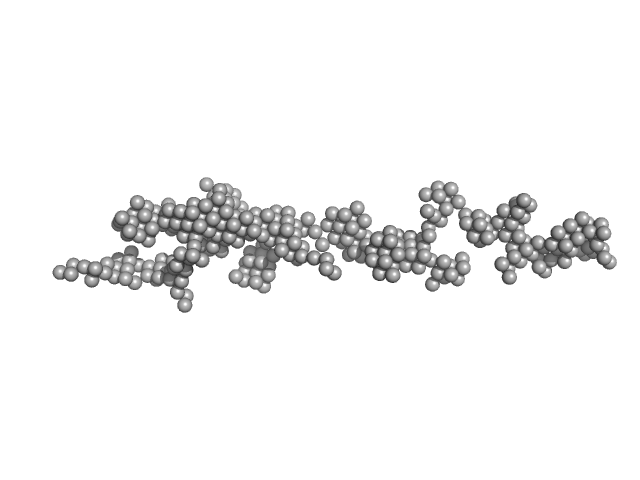
|
| Sample: |
Amyloid-beta precursor protein, 5 kDa Homo sapiens protein
|
| Buffer: |
1 mM Hepes, 0.12 % NH4OH, pH: 10.7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Apr 11
|
Time-resolved small-angle X-ray scattering study of polymorphic nature of Aß42 oligomerisation
Tia Cheremnykh
|
| RgGuinier |
5.8 |
nm |
| Dmax |
42.0 |
nm |
|
|
UniProt ID: P05067 (672-713) Amyloid-beta precursor protein
|
|
|

|
| Sample: |
Amyloid-beta precursor protein, 5 kDa Homo sapiens protein
|
| Buffer: |
1 mM Hepes, 0.12 % NH4OH, pH: 10.7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Dec 13
|
Time-resolved small-angle X-ray scattering study of polymorphic nature of Aß42 oligomerisation
Tia Cheremnykh
|
| RgGuinier |
6.9 |
nm |
| Dmax |
60.0 |
nm |
|
|
UniProt ID: P05067 (672-713) Amyloid-beta precursor protein
|
|
|

|
| Sample: |
Amyloid-beta precursor protein, 5 kDa Homo sapiens protein
|
| Buffer: |
1 mM Hepes, 0.1 % NH4OH (pH∼10.7), pH: 10.7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Oct 31
|
Time-resolved small-angle X-ray scattering study of polymorphic nature of Aß42 oligomerisation
Tia Cheremnykh
|
| RgGuinier |
2.5 |
nm |
| Dmax |
6.5 |
nm |
|
|
UniProt ID: P05067 (672-713) Amyloid-beta precursor protein
|
|
|
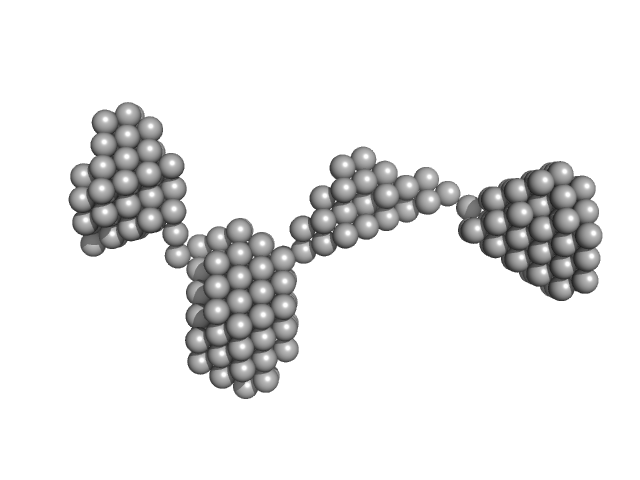
|
| Sample: |
Amyloid-beta precursor protein, 5 kDa Homo sapiens protein
|
| Buffer: |
1 mM Hepes, 0.1 % NH4OH (pH∼10.7), pH: 10.7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Nov 1
|
Time-resolved small-angle X-ray scattering study of polymorphic nature of Aß42 oligomerisation
Tia Cheremnykh
|
| RgGuinier |
3.7 |
nm |
| Dmax |
9.1 |
nm |
|
|
UniProt ID: P05067 (672-713) Amyloid-beta precursor protein
|
|
|
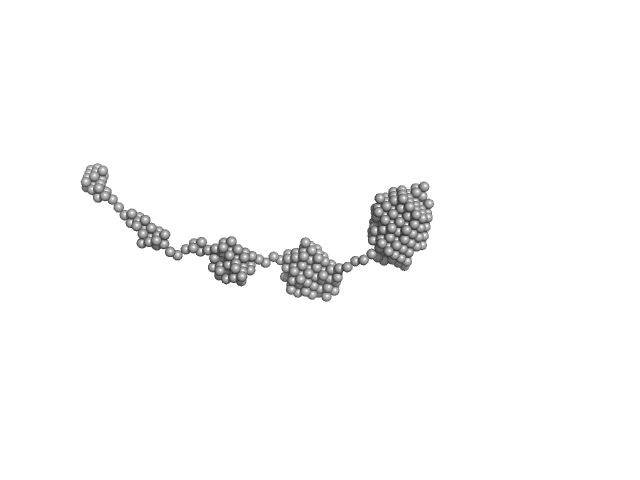
|
| Sample: |
Amyloid-beta precursor protein, 5 kDa Homo sapiens protein
|
| Buffer: |
1 mM Hepes, 0.1 % NH4OH (pH∼10.7), pH: 10.7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Nov 1
|
Time-resolved small-angle X-ray scattering study of polymorphic nature of Aß42 oligomerisation
Tia Cheremnykh
|
| RgGuinier |
3.7 |
nm |
| Dmax |
11.0 |
nm |
|
|